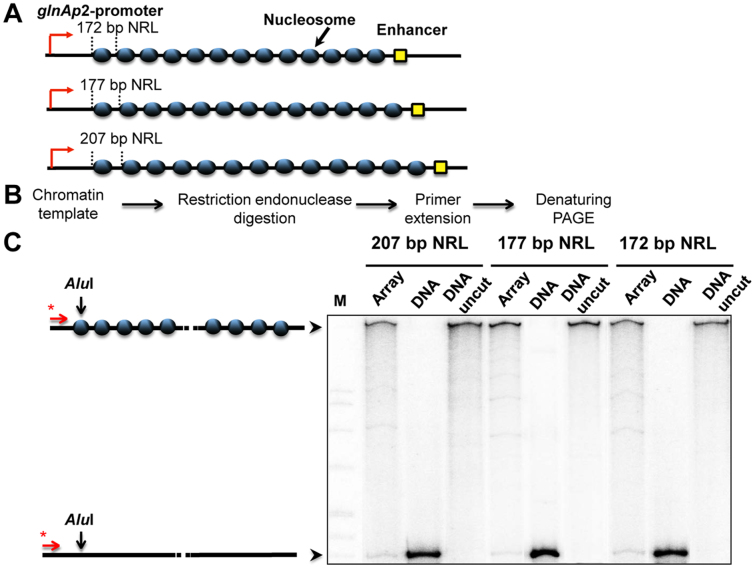
Figure 1.
Analysis of nucleosome occupancy of 601 arrays having different nucleosome repeat lengths. (A) Schematic diagram of 13-nucleosome arrays having 172-, 177- and 207-bp nucleosomal repeat lengths (NRL). (B) Experimental approach: a restriction enzyme sensitivity assay. 601207×13, nucleosomal arrays were assembled on linearized plasmids at different ratios of histones to DNA and incubated in the presence of an excess of restriction enzyme AluI (see Materials and Methods). Purified DNA was subjected to primer extension followed by denaturing PAGE. (C) Saturated 13-nucleosomal arrays (207-, 177- and 172-bp NRL) were characterized using the restriction digestion sensitivity assay. Analysis of end-labeled DNA by denaturing PAGE (right). Note that chromatin assembly results in almost quantitative protection of the templates from AluI. M: MspI digest of plasmid pBR322 (left).