Figure 6.
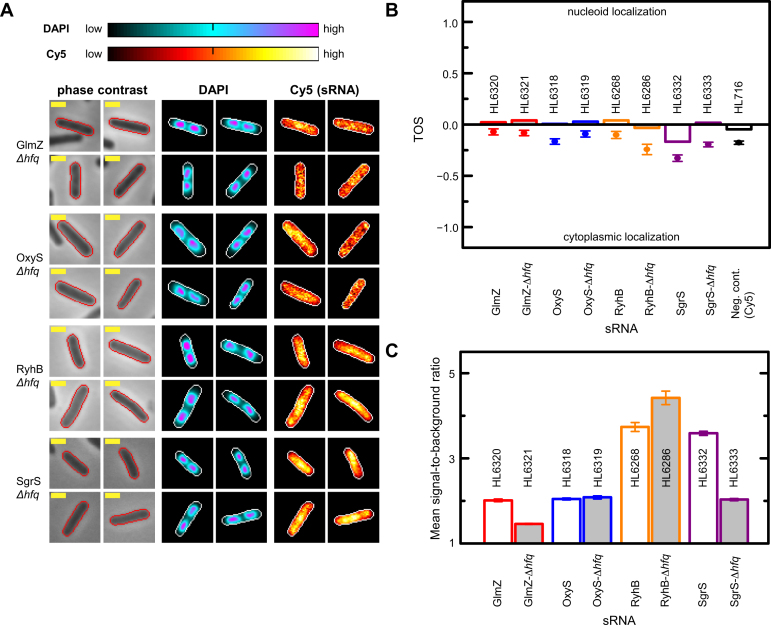
Hfq has minimal effect on sRNA localization. Cell edges were identified as in Figure 2A. Yellow scale bar indicates 1 μm for all images. Measurements for the negative control strain (HL716) without any plasmid and probes were made at 1 mM IPTG. (A) sRNA localization in strains without hfq (Δhfq). Signal intensities of DAPI and Cy5 in individual cells are shown as heat maps with cellular normalization. All strains have the sRNA transcribed from the PCon promoter. Strains: glmZ (HL6321; n = 144); oxyS (HL6319; n = 97); ryhB (HL6286; n = 62); and sgrS (HL6333; n = 326). (B) TOS for each sRNA with hfq (data from the experiments in Figure 2A) and without hfq (panel A). Bars are the medians, circle symbols are the means and error bars are the SEMs. Median TOS for pairs of strains with or without hfq were very similar for GlmZ, OxyS, and RyhB (P = 4.4 × 10−1, 1.3 × 10−2 and 1.7 × 10−2 respectively; Mann–Whitney U two-tailed test). Median TOS was significantly different for SgrS with or without hfq (P = 5.2 × 10−4). (C) Signal-to-background ratios for each sRNA with and without hfq. Error bars are the SEMs. Mean signal-to-background ratio in strains with and without hfq were statistically significant for GlmZ, RyhB and SgrS but not for OxyS (P = 1.4 × 10−50, 2.6 × 10−4, 4.0 × 10−125 and 3.7 × 10−1 respectively; two-tailed t-test).