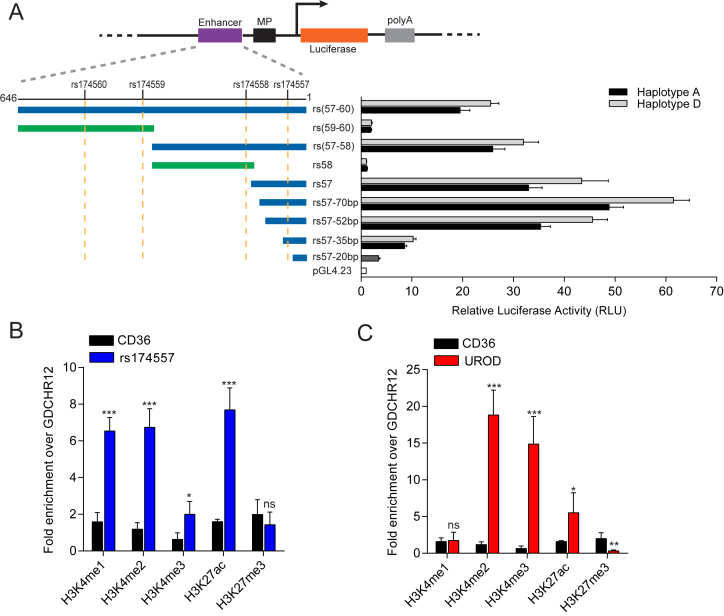
Figure 1.
Identification of rs174557 as the functional variant. (A) Only luciferase constructs harboring rs174557 showed strong enhancer activity with the minimal enhancer region mapped to fragment rs57-70bp. Fragments containing each haplotype or allele were inserted upstream of the minimal promoter sequence of pGL4.23. The luciferase activities were normalized to the empty control vector pGL4.23. All the constructs containing rs174557 showed significant differences (P < 0.0001) between haplotype D (light gray bars) and haplotype A (black bars) in luciferase assay. Values are expressed as the mean from three independent transfections each with six technical replicates. Error bars represent SD from all technical replicates. Abbreviations are as follows: MP, minimal promoter; and polyA, the SV40 polyA signal. (B) The rs174557 locus showed classical enhancer features in HepG2 cells identified by ChIP-qPCR on different histone modifications. (C) The transcription start site of UROD gene was employed as the positive control for ChIP. For B and C, two genomic regions CD36 and GDCHR12 were used as negative controls. All the values are expressed as fold changes over GDCHR12. The error bars represent SD from five replicates with P value calculated by two-tailed t tests. *P < 0.05; **P < 0.01; ***P < 0.001 and ns, not significant compared with GDCHR12.