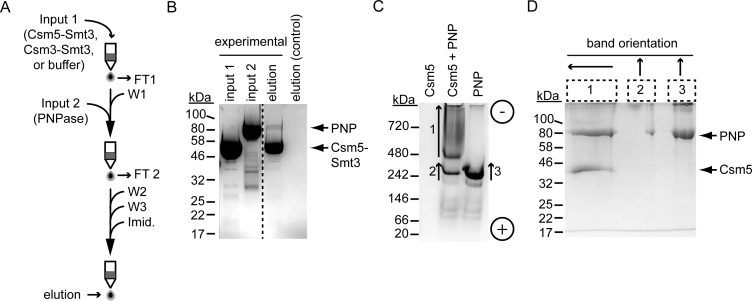
Figure 5.
Csm5 physically interacts with PNPase. (A) Abbreviated experimental flow of a Ni2+ affinity pulldown assay, in which His10Smt3 tagged Csm5 (Csm5-Smt3, ‘input 1’, 1 nmol) is loaded onto a column containing Ni2+-agarose beads and washed. Untagged PNPase (‘input 2’, 0.7 nmol) is then passed through the column, and anything unbound is thoroughly washed. Proteins retained in the column are eluted using 500 mM imidazole. Abbreviations are as follows: FT, flow-through; W, wash; imid., imidazole. (B) Samples from the Csm5–Smt3 (input 1), untagged PNPase (input 2), and final elution were resolved using SDS-PAGE. The final elution from a negative control is also shown in which dialysis buffer was used as input 1. See Supplementary Figure S5 for a detailed description of the experiment and all samples collected during the experiment. The dotted line separates non-contiguous lanes within the same gel. Shown is a representative of four independent trials. (C) Csm5 and PNPase (100 pmol each) were resolved on a 5% vertical native gel alone or in combination (1:1 ratio). The position of (+) and (-) electrodes are indicated. (D) Numbered bands shown in panel C were excised from the native gel and the proteins within were resolved using denaturing SDS-PAGE. Arrowheads mark the top of each excised band and indicate band orientation when run in second dimension. Shown is a representative of three independent trials. Proteins were visualized with Coomassie G-250.