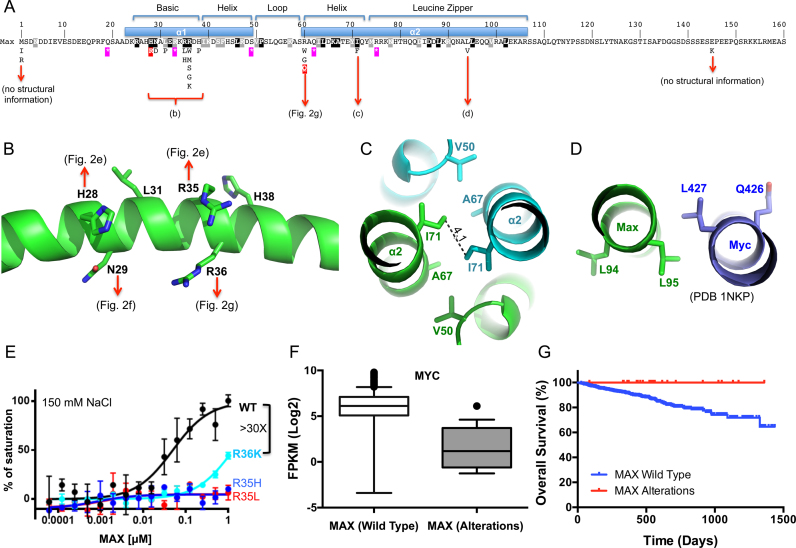
Figure 5.
MAX alterations in multiple myeloma. (A) Relative position of MAX variants detected in multiple myeloma and their relationship to the structural features of the MAX protein. Features of the DNA binding (bHLH) and dimerization domains (leucine zipper) are shown. Missense mutations are shown below. Purple stars show the position of nonsense mutations. Black/gray shaded residues are those conserved across the MAX bHLH family (see Supplementary Figure S3D). Those ‘hotspot’ mutations that were biochemically analyzed (H28R, R60Q) are marked in red. (B–D) Structural impacts of MAX variants in different regions of the MAX protein. (B) The detailed impact of missense mutations in the basic region—detailed interactions can be observed in panels of Figure 2E, F and G. (C) An I71 to F variant is likely to interfere with the dimer interaction via the leucine zipper. (D) The L94V, located on the outer surface of helix α2, has the potential to alter interactions with regions outside of structured region of the dimer or other proteins. (E) R36K reduced DNA binding significantly, whereas the R35H and R35L completely abolished binding under the same conditions. Data represent two independent determinations performed in duplicate. (F) Comparison of MYC expression in multiple myeloma samples from patients with wild type MAX (n = 625) or MAX alterations (n = 18). Shown are box plots of the MYC expression levels between samples in each class. Median is indicated by a line, box is the 1st and 3rd quartiles, whiskers are the most extreme data point that is 1.5 times the interquartile range. Significance (P < 0.0001) was assessed by the Mann–Whitney U test. (G) Survival outcomes among Multiple Myeloma patients with wild-type MAX (n = 740) or MAX alterations (n = 24). Patients with MAX alterations exhibit a significantly better outcomes [Mantel–Cox log rank test, P = 0.041 (95% CI, 0.13–0.96)].