Abstract
Arabidopsis, miR402 that is encoded within the first intron of a protein-coding gene At1g77230, is induced by heat stress. Its upregulation correlates with splicing inhibition and intronic proximal polyA site selection. It suggests that miR402 is not processed from an intron, but rather from a shorter transcript after selection of the proximal polyA site within this intron. Recently, introns and active 5΄ splice sites (5΄ss’) have been shown to stimulate the accumulation of miRNAs encoded within the first exons of intron-containing MIR genes. In contrast, we have observed the opposite effect of splicing inhibition on intronic miR402 production. Transient expression experiments performed in tobacco leaves revealed a significant accumulation of the intronic mature miR402 when the 5΄ss of the miR402-hosting intron was inactivated. In contrast, when the miR402 stem-loop structure was moved into the first exon, mutation of the first-intron 5΄ss resulted in a decrease in the miRNA level. Thus, the 5΄ss controls the efficiency of miRNA biogenesis. We also show that the SERRATE protein (a key component of the plant microprocessor) colocalizes and interacts with several U1 snRNP auxiliary proteins. We postulate that SERRATE-spliceosome connections have a direct effect on miRNA maturation.
INTRODUCTION
MicroRNAs (miRNAs) are short, single stranded, non-coding RNAs that act at the post-transcriptional level leading to mRNA cleavage and degradation or translational repression by base pairing to their target mRNAs (1–3). These small RNAs can be considered a quick reaction force that allows rapid developmental changes and responses to environmental cues (4–10). Consequently, prompt and efficient miRNA action requires the existence of many regulatory mechanisms that fine-tune their levels in response to cellular needs. However, the mechanism of plant miRNA production still remains unclear.
In contrast to mammalian cells, plant miRNA biogenesis occurs primarily in the nucleus (11). Plant primary transcripts of MIR genes (pri-miRNAs) are synthesized by RNA polymerase II and thus contain caps and polyA tails (12). Processing of pri-miRNA to mature miRNA is performed by an RNase III ribonuclease, DICER-LIKE1 (DCL1), which is assisted primarily by HYPONASTIC LEAVES 1 (HYL1) and SERRATE (SE), the protein involved in miRNA biogenesis and pre-mRNA splicing (13–17). Recently, additional players involved in miRNA maturation have been discovered, from which several affect miRNA accumulation additionally via regulation of pri-miRNA splicing efficiency (18–26). These are: HOS5, RS40/RS41, STA1 and GRP7 (18,27,28). Thus, the structure of MIR genes is an important factor that facilitates the complex regulation of miRNA production (29–33).
In accordance with the latest scientific reports, from 75 Arabidopsis thaliana MIR gene structures characterized thus far, 46 encode independent transcription units and approximately half of them contain introns (34–36). Another 29 miRNAs are located within the introns of other genes that encode protein-coding or non-coding RNAs (37,38). Both pri-miRNAs and miRNA-hosting pre-mRNAs undergo constitutive and alternative splicing. Moreover, many alternative polyadenylation signals have been identified in Arabidopsis pri-miRNAs (35,36). In fact, these post-trascriptional events have been already reported to regulate miRNA biogenesis. For exonic MIR161, MIR163 and MIR172a pri-miRNAs (each containing one intron), splicing stimulates mature miRNA production and the recognition of active 5΄ splice sites (5΄ss) (located downstream of the miRNA stem-loop structure) by U1 snRNP is crucial for the stimulation of miRNA biogenesis (29,30). These results suggest the existence of communication between the spliceosome and the plant microprocessor in the case of miRNAs located within the upstream exons of intron-containing genes. However, the key players in this crosstalk and the effects of these interactions on miRNAs present in introns remain unknown.
Reports concerning intronic miRNAs focus primarily on the mechanism of their biogenesis in humans and animals. Knockdown of a U1 snRNP-specific protein reduced the level of miR211 (39). In addition, the production of miR25, miR93 and miR106b was significantly affected by non-canonical splicing events, alternative transcription start site and polyadenylation site selection (40,41). These results suggest that intronic miRNA maturation, at least in human and animal cells, can be regulated at different levels of transcriptional and co-transcriptional processing of their host pre-mRNAs and pri-miRNAs.
In contrast, information regarding intronic miRNA biogenesis in plants is scarce. In 2008, possible mechanisms of intronic miRNA biogenesis were proposed that assumed either cooperation or competition between the plant microprocessor and the spliceosome (37). The later model of intronic miR400 maturation assumed that heat stress activation of the alternative 5΄ss, which was downstream of the miRNA stem-loop structure, resulted in lower levels of mature miRNA (38). This observation posited that plant intronic miRNAs are produced efficiently from spliced-out introns. Another specific class of miRNAs produced from introns are mirtrons, of which the maturation in animals does not require the nuclear step of miRNA biogenesis; instead, their precursors are products of spliceosome activity. In plants, 5 and 18 mirtron-like intervening sequences have been identified thus far in A. thaliana and Oryza sativa, respectively, using bioinformatics tools (42,43). However, the mechanism of their maturation comprising the involvement of the spliceosome and the microprocessor has still yet to be uncovered. The complex structure of plant MIR genes suggests that intronic miRNA biogenesis is affected by the interplay between different RNA metabolism machineries already at the primary transcript level similar to that in animals.
The results presented in this study show that SE, a key component of the plant microprocessor interacts with U1 snRNP auxiliary proteins, supporting the importance of SE–spliceosome communication in the regulation of miRNA biogenesis. For at least two intronic miRNAs, splicing inhibition lead to strong upregulation of their levels. Interestingly, we discovered that the translocation of the intronic miR402 stem-loop structure into the upstream exon downregulated miRNA production when the 5΄ss was inactivated. These results show that the position of the miRNA hairpin with regard to the 5΄ss strongly affects miRNA maturation. The presented results show novel mechanisms of regulating plant miRNA biogenesis and target mRNA level.
MATERIALS AND METHODS
Plant material
Arabidopsis thaliana (wild-type, se-1 and se-2 (44,45) seeds were stratified on solid  MS (Murashige and Skoog, Sigma-Aldrich) medium supplemented with 1% agar (Sigma-Aldrich) for 3 days at 4°C and then transferred to 22°C. Seedlings were grown for 14 days (16/8 h light/dark, 70% humidity, 150–200 μmol m-2 s-1 photon flux density).
MS (Murashige and Skoog, Sigma-Aldrich) medium supplemented with 1% agar (Sigma-Aldrich) for 3 days at 4°C and then transferred to 22°C. Seedlings were grown for 14 days (16/8 h light/dark, 70% humidity, 150–200 μmol m-2 s-1 photon flux density).
For heat treatment, 2-week-old seedlings were exposed to 37°C and plants were collected at the indicated times. Plant material for the abiotic stress analyses was prepared as previously described (9).
The half-life of pri-miR402 was determined as previously described (46) with the following modifications. Arabidopsis 17-day-old seedlings grown on solid 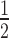 MS medium were exposed to 37°C for 2 h, then transferred into incubation buffer (1 mM Pipes pH 6.25, 1 mM sodium citrate, 1 mM KCl, 15 mM sucrose) and swirled at 30 rpm at 22°C for 30 min. Next, 3΄-deoxyadenosine (cordycepin; C3394; Sigma Aldrich) was added to a final concentration of 0.6 mM (time 0) and tissue samples were collected after 20, 40, 60 and 80 min of incubation.
MS medium were exposed to 37°C for 2 h, then transferred into incubation buffer (1 mM Pipes pH 6.25, 1 mM sodium citrate, 1 mM KCl, 15 mM sucrose) and swirled at 30 rpm at 22°C for 30 min. Next, 3΄-deoxyadenosine (cordycepin; C3394; Sigma Aldrich) was added to a final concentration of 0.6 mM (time 0) and tissue samples were collected after 20, 40, 60 and 80 min of incubation.
To obtain 35-day-old A. thaliana Col-0 wild-type plants, seeds after 3-day stratification were grown at 22°C (16/8 h light/dark, 50–60% humidity, 150–200 μmol m-2 s-1 photon flux density) in Jiffy-7 pots (Jiffy).
Nicotiana benthamiana plants were grown for 6 weeks at 25°C (16/8 h light/dark, 50% humidity, 150–200 μmol m-2 s-1 photon flux density).
Construct preparation
For miR402-hosting gene construct preparation, the genomic sequence of At1g77230 was amplified (from genomic DNA isolated using a DNeasy Plant Mini Kit, Qiagen) and cloned into the pENTR/D-TOPO plasmid (ThermoFisher Scientific) using NotI and AscI restriction sites. The mutagenesis of identified splice sites was performed using a QuikChange® II Site-Directed Mutagenesis Kit (Agilent Technologies). To obtain expression in plants, At1g77230 gene versions were cloned into the pMDC32 Gateway binary vector (47) using Gateway LR Clonase II Enzyme Mix (ThermoFisher Scientific).
For the promoter influence analysis, the modified pMDC99 Gateway binary vectors (47) were prepared, carrying native At1g77230, ACT2 (At3g8780) and GAPDH (At1g13440) promoter regions, representing sequences located ∼1500, ∼2000 and ∼2000 bp upstream of the At1g77230, At3g8780 and At1g13440 TSSs, respectively. The amplified fragments were cloned upstream of the pMDC99 attR1 sequence using the combination of HindIII and AscI restriction sites. Next, the At1g77230 gene versions with active (native) and inactivated (Δ5΄ss) the first-intron constitutive 5΄ss were cloned into prepared binary vectors using Gateway LR Clonase II Enzyme Mix (ThermoFisher Scientific).
To change the location of the miR402 stem-loop structure, the hairpin was removed using a three-step polymerase chain reaction (PCR) approach (step 1: amplification of the 1–400 nt fragment; step 2: amplification of the 710–3649 nt fragment; step 3: amplification of the 1–3649 nt fragment based on a template consisting of the mixed PCR products from steps 1 and 2). Then, pre-miR402 was amplified independently using specific primers. Next, the miR402 stem-loop structure was cloned into the first exon using the MfeI restriction site. To insert the pre-miR402 into the second intron, the three-step PCR approach was used based on the template without the miR402 hairpin (step 1: amplification of the 1–1211 nt fragment; 2: amplification of the 1212–3649 nt fragment; step 3: amplification of the 1–3649 nt fragment based on a template consisting of the pre-miR402 and PCR products of steps 1 and 2). To obtain expression in plants, At1g77230 gene versions were cloned into the pMDC32 Gateway binary vector (47) using Gateway LR Clonase II Enzyme Mix (ThermoFisher Scientific).
To analyze protein interactions in yeast cells, pGADT7 (with a Gal4 activation domain) and pGBKT7 (with a Gal4 binding domain) cloning vectors were used (Clontech). The coding sequences (CDS) of SE (At2g27100), AtU1A (At2g47580), AtU1C (At4g03120), AtU1-70K (At3g50670), AtPRP39a (At1g04080), AtPRP39b (At5g46400), AtPRP40a (At1g44910), AtPRP40b (At3g19670), AtLUC7a (At3g03340), AtLUC7b (At5g17440) and AtLUC7rl (At5g51410) were amplified using cDNA template prepared from 35-day-old rosette leaves.
For protein pull-down assay constructs encoding the MBP-SE, MBP-GFP and Tetratricopeptide repeat (TPR) domain of SGT1 were used (17). AtPRP39b, AtPRP40a, AtPRP40b and AtLUC7rl coding sequences cloned into pGBKT7 were used as the templates for in vitro translation.
For microscopic analyses, coding sequences of the studied proteins (all mentioned above and AtCBP20 (At5g44200)) were first cloned into pENTR/D-TOPO (ThermoFisher Scientific) using topoisomerase I-based 5-min directional ligation or NotI and AscI restriction sites. Next, Gateway LR Clonase II Enzyme Mix (ThermoFisher Scientific) was used to generate expression clones based on destination plasmids with the GFP or the tagRFP sequence under control of the UBQ10 promoter. For constructs mimicking SE in the se-2 and se-1 mutants, stop codons were inserted after nucleotide 2043 (using a QuikChange® II Site-Directed Mutagenesis Kit; Agilent Technologies) and nucleotide 2103 of SE CDS, respectively.
All of the constructs and mutations were confirmed by DNA sequencing. The oligonucleotide sequences are listed in Supplementary Table S1.
Transient expression in N. benthamiana leaves
Agroinfiltration was performed as previously described (29). For the determination of pri-miR402 half-life 72 hours after agroinfiltration small discs (5 mm diameter) were collected (n = 100) and placed in a square Petri dish containing 30 ml of incubation buffer (1 mM Pipes pH 6.25, 1 mM sodium citrate, 1 mM KCl, 15 mM sucrose). After 30 min of swirling at 30 rpm 3΄-deoxyadenosine (cordycepin; C3394; Sigma Aldrich) was added to a final concentration of 0.6 mM (time 0), and tissue samples were collected after 30, 60, 120 and 180 min of incubation.
RNA isolation and cDNA preparation
Total RNA for cDNA preparation was isolated using a Direct-zol™ RNA Mini Prep Kit (Zymo Research) and treated with Turbo DNase I (ThermoFisher Scientific). For analyses of the At1g77230 transcript expression levels, RT reactions were prepared with the use of SuperScript III Reverse Transcriptase (ThermoFisher Scientific), oligo-dT(18) primer (ThermoFisher Scientific) and 1 μg (for miR402 transcript half-life measurements) or 3 μg of total DNase-treated RNA.
For mature miRNA analyses, RT reactions were prepared using MultiScribe™ Reverse Transcriptase (ThermoFisher Scientific), 10 ng of total DNase-treated RNA and 5× RT primer specific for each miRNA or U6 snRNA used as a reference (ThermoFisher Scientific).
RT-qPCR
RT-qPCR was performed with Power SYBR® Green PCR Master Mix (ThermoFisher Scientific) using a 7900HT Fast Real-Time PCR System (ThermoFisher Scientific) as previously described (48). The amplification efficiency of each primer pair (final concentration, 200 nM each) was calculated by making 10-fold dilution series of the mix of templates, calculating a linear regression based on the data points and estimating the efficiency from the line slope. In addition, the dissociation curve was always analyzed to exclude the possibility of multiple products amplification. Only primer pairs with the highest, almost equal efficiency (max. difference of 2% was approved) and with only one peak visible on the dissociation curve were used for analysis. To avoid any ambiguity the RT-qPCR products were sequenced to confirm that only the isoform of interest was amplified.
Expression levels were calculated using the relative quantification (2-ΔCt), while the fold change was calculated using the 2-ΔΔCt method. The mRNA fragments of glyceraldehyde-3-phosphate dehydrogenase (GAPDH, At1g13440), and HYGROMYCIN (ACI22368) were amplified and detected simultaneously as reference genes for A. thaliana and N. benthamiana, respectively. To estimate the splicing efficiency, all events identified within the analyzed intron were summed and treated as 100%, and the contribution of fully spliced and unspliced isoforms was calculated. To determine statistical significance each individual isoform was analyzed in the sample of interest against the control sample (or other relevant sample as indicated in the figure legend). To calculate the level of At1g77230 transcripts terminated at the proximal polyA site, the results obtained with the use of primers anchored to the first exon of At1g77230 were diminished by the expression level of transcripts ended at the distal polyA sites. Next the ratio of polyA site selection was calculated and statistical significance was determined for each individual isoform against the control sample. For evaluation of the pri-miR402 stability the levels of the analyzed transcripts were plotted against time. Next, the linear regression was applied to calculate the half-life of transcripts.
Each RT-qPCR was performed independently for at least three biological replicates. All results were analyzed using SDS 2.4 software (ThermoFisher Scientific). Error bars were calculated using the SD Function in Microsoft Excel software. The statistical significance of the results presented was estimated using Student's t-test or ANOVA followed by the Tukey's test (for the studies of an influence of se-1 and se-2 on miRNA biogenesis) at three significance levels: *P < 0.05, **P < 0.01 and ***P < 0.001.
RT-qPCR analyses of mature miRNA expression levels were performed according to the TaqMan® MicroRNA & Non-coding RNA Assay (ThermoFisher Scientific) using TaqMan® Universal Master Mix II with UNG (ThermoFisher Scientific), TaqMan® probes and primers specific for mature miRNAs and internal reference genes (U6 snRNA (At3g14735) and HYGROMYCIN (ACI22368)) for A. thaliana and N. benthamiana, respectively.
All oligonucleotide sequences used for RT-qPCR experiments are listed in Supplementary Table S1.
3΄ RACE
Total RNA isolated with the use of a Direct-zol™ RNA Mini Prep Kit (Zymo Research) was treated with Turbo DNase I (ThermoFisher Scientific) and used for cDNA template preparation according to the manufacturer's instructions (SMARTer RACE cDNA Amplification Kit (Clontech)). PCR reactions were performed using the Advantage 2 PCR Enzyme System (Clontech). The primer sequences are listed in Supplementary Table S1. Next, the PCR products were cloned into the pGEM T-Easy vector (Promega) and sequenced. Identified alternative polyadenylation sites are listed in Table 1.
Table 1. Identified alternative polyadenylation sites of At1g77230 transcripts.
| No. | Name | Position (from the 5΄ end of At1g77230 transcript) in nucleotides | Sequence of the identified polyadenylation cleavage site (bold) |
|---|---|---|---|
| 1 | proximal polyA site | 974 | AAGTGAATT |
| 2 | distal polyA site 1 | 1756 | TAATGAACC |
| 3 | distal polyA site 2 | 1889 | TTTCCAGTA |
RNA isolation and northern hybridization of mature miR402
Total RNA was isolated with TRIzol reagent (ThermoFisher Scientific). Northern experiments were performed as previously described (31).
Yeast two-hybrid analysis
The yeast two-hybrid (YTH) experiments were performed using the Matchmaker GAL4-based two-hybrid system (Clontech) according to the manufacturer's protocol. Saccharomyces cerevisiae strain Y2HGold was cotransformed with 700 ng of each plasmid encoding BD- and AD-fusion proteins according to the manufacturer's protocol, and spread on an SD/-Leu/-Trp selective medium (-LT). To test for protein interactions, the cells were then transferred to SD-Leu-Trp-His-Ade (-LTHA) and SD-Leu-Trp-His-Ade+X-α-Gal+Aureobasidin A (-LTHA+X-α-Gal+Aur) plates and grown for three days at 28°C. The experiments were performed in three replicates.
Protein pull-down assay
Assays were performed as previously described (17). The overexpression of SE and GFP (as a negative control) fused with maltose-binding protein (MBP) was performed in Escherichia coli strain BL21(DE3)RIL as follows: cells were grown for 16 h at 20°C (MBP-SE) or 2 h at 37°C (MBP-GFP) after induction by 0.4 mM isopropyl β-D-1-thiogalactopyranoside, then harvested and sonicated (15 cycles of 30 s ON and 30 s OFF using a Bioruptor Plus, Diagenode) in MBP buffer (20 mM Tris–HCl, 0.2 M NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA) and Complete Mini EDTA-free protease inhibitor tablets (Roche), pH 7.4). After sonication, lysates were centrifuged for 15 min at 14 000 × g at 4°C and the supernatants containing the protein extract were collected. AtPRP39b, AtPRP40a, AtPRP40b, AtLUC7rl and TPR domain of the SGT1 protein were obtained by in vitro translation (TNT T7 Coupled Wheat Germ Extract System (Promega)) in the presence of [35S]-methionine (Hartmann Analytic GmbH).
For pull-down experiments, MBP-SE and MBP-GFP from bacterial lysates were bound to amylose beads (New England Biolabs), washed three times with MBP buffer (20 mM Tris–HCl, 0.2 mM NaCl, 1 mM EDTA and Complete Mini EDTA-free protease inhibitors tablets (Roche), pH 7.4) and incubated with labeled AtPRP39b, AtPRP40a, AtPRP40b, AtLUC7rl and TPR domain of SGT1 in phosphate buffer (28 mM NaH2PO4, 72 mM Na2HPO4, 250 mM KCl and 0.5% Triton X-100) for 2 h at 4°C. Next, the beads were washed four times with phosphate buffer and the protein complexes were eluted with 10 mM maltose. The labeled proteins were separated in 14% sodium dodecyl sulphate-polyacrylamide gel and detected with an image analyzer (FLA-5000 (Fujifilm)). To verify that an equivalent amount of the MBP-SE and MBP-GFP proteins was bound to the beads, samples of the MBP proteins bound on the amylose beads were diluted hundred times and Western blot was performed with the use of anti-MBP (Sigma-Aldrich, M6295, 1:10 000) and anti-rat (Merck Millipore, AP136P, 1:10 000) antibodies.
Mesophyll protoplast transfection
The fusion constructs used for protein localization and FRET-FLIM analyses were introduced into A. thaliana wild-type Col-0 protoplasts isolated from rosette leaves of plants grown for 3–4 weeks at 20–22°C (16/8 h light/dark, 50–60% humidity, 150–200 μmol m-2 s-1 photon flux density). Leaves prepared by the Tape-Arabidopsis Sandwich method (49) were soaked in 20 ml of the enzyme solution (1% (w/v) Cellulase Onozuka R10 from Trichoderma viride (Serva; 1 U/mg) and 0.2% (w/v) Macerozyme R-10 from Rhizopus sp. (Serva) dissolved in mannitol solution (0.4 M mannitol, 20 mM KCl and 20 mM 2-[N-morpholino]ethanesulfonic acid (MES) pH 5.7)), supplemented with CaCl2 to a final concentration of 10 mM in a Petri dish and incubated on a platform shaker (40-60 rpm) for 30 min at room temperature (50). Protoplasts were then collected by centrifugation for 3 min at 200 × g, washed three times in W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl and 2 mM MES pH 5.7) and resuspended in 1.2 ml of MMg solution (0.2 M mannitol, 15 mM MgCl2 and 4 mM MES pH 5.7).
Samples (200 μl) of the protoplast suspension were transferred into round-bottom tubes containing 5 μg of plasmid DNA (5 μg of each plasmid in the case of cotransfection), and 220 μl of PEG/Ca2+ solution (40% (w/v) polyethylene glycol 4000 (PEG, Sigma-Aldrich), 0.2 M mannitol and 100 mM Ca(NO3)2) was then added. The suspension was mixed well and incubated for 5 min at room temperature, and 800 μl of W5 solution was added to stop the transfection process. The protoplasts were collected by centrifugation for 3 min at 200 × g, washed in W5 solution, resuspended in 1 ml of W5 solution containing 1 mM glucose and incubated for 7 h in the dark at 22°C before imaging and FRET-FLIM measurements.
Imaging
The subcellular localization of proteins was examined with a confocal scanning microscope (Nikon A1Rsi) using 40×/1.25 and 60×/1.20 water-immersion objectives. Excitation was achieved with an argon laser at 488 nm (GFP) and with a diode laser at 561 nm (tagRFP). Fluorescence was observed using the emission spectrum range of 525/50 nm (GFP) and 595/50 (tagRFP). Images were prepared using Fiji software (51).
FRET-FLIM
FRET-FLIM was performed on a PicoHarp300-Dual Channel SPAD system (PicoQuant) in combination with Nikon A1Rsi microscope and on Leica TCS SP5 X scan head coupled to internal Hybrid (HyD) detector (Becker & Hickl). The excitation pulses were generated by a Picosecond Pulsed Diode Laser LDH-D-C-485 (PicoQuant) and supercontinuum tunable white light laser (488 nm), resulting in excitation pulses of 100 ps at a repetition of 40 MHz. 60×/1.20 water-immersion and 63×/1.20 water-immersion objectives were used. Images with a frame size of 128 × 128 pixels were acquired, and the average count rate was around 104 photons per second for an acquisition time of ±60 s. From the time resolved fluorescence intensity images, the fluorescence decay curves were calculated per pixel and fitted with double-exponential decay model using the SymPhoTime 64 (PicoQuant) and the SPCImage 3.10 software (Becker &Hickl). The fitting procedure was performed without fixing any parameters. Several cells (n > 10) for each sample were analyzed and Student's t-test was used to determine statistical significance of differences between samples (*P < 0.001). Error bars represent standard error of the mean (SEM) (52).
RESULTS
Stress-induced miR402 accumulation correlates with splicing inhibition of the miRNA-hosting intron and proximal polyadenylation site selection
To reveal the mechanism of plant intronic miRNA biogenesis, we selected miR402, which is located within the first intron of the At1g77230 gene encoding a tetratricopeptide repeat-rich protein of unknown function (Figure 1A). Mature miR402 has been shown to be responsive to many environmental cues such as cold, heat, salinity, ABA treatment and dehydration (7,53). These findings prompted us to perform a deeper analysis of miR402 biogenesis regulation under different stress conditions.
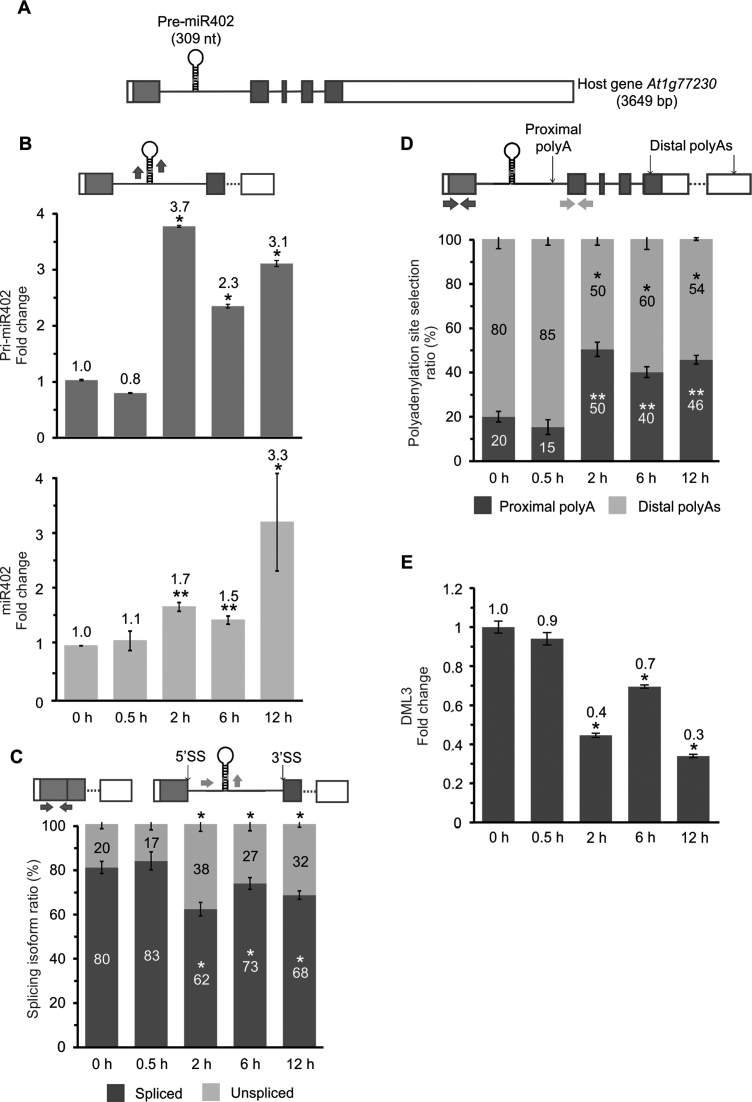
Figure 1.
Heat-induced miR402 accumulation correlates with the inhibition of miRNA-carrying intron splicing and higher selection of proximal polyadenylation site. (A) Structure of At1g77230, hosting pre-miR402 within the first intron. Boxes represent exons (protein-coding sequence marked in gray, UTRs in white), and lines depict introns. RT-qPCR analysis of (B) the pri-miR402 (upper panel) and miR402 (lower panel) levels, (C) miR402-carrying intron splicing efficiency, (D) the proximal and distal polyA site usage and (E) DML3 expression levels in control and heat stress conditions. Arrows in the upper parts of the panels with the At1g77230 gene scheme depict the primers used (B–D). Error bars indicate SD (n = 3), and the asterisk indicates a significant difference between the sample and control (*P < 0.05; **P < 0.01; white stars—for spliced or proximal polyA isoforms; black stars—for unspliced and distal polyA isoforms (C-D)).
First, the effect of heat stress on miR402 maturation was confirmed. A. thaliana 2-week-old seedlings were subjected to 37°C for 0.5, 2, 6 and 12 h. RT-qPCR analysis showed upregulation of pri-miR402 and mature miR402 levels after at least 2 h of heat treatment (Figure 1B and Supplementary Figure S1A). We also discovered that high temperature inhibited the splicing of the miR402-carrying intron (Figure 1C), but we observed no changes of the splicing of the arbitrarily selected At1g77230 intron no. four that does not carry any miRNA (Supplementary Figure S1B). Thus, heat stress clearly does not affect the splicing efficiency of all introns of the miR402 host gene. Furthermore, we identified three alternative polyA sites in At1g77230 transcripts in the following regions using the 3΄ RACE approach: (i) within the first intron downstream of pre-miR402 (subsequently called the proximal polyA site), and (ii) in the last exon (two polyA sites subsequently called distal polyA sites) (Table 1). These polyA sites were also found in Direct RNA Sequencing data provided by others (54). Interestingly, the use of the proximal polyA site increased significantly upon heat stress (Figure 1D). Additionally, miR402 is known to target the mRNA of DEMETER-LIKE PROTEIN 3 (DML3, At4g34060) (55). RT-qPCR analyses of the DML3 transcript level showed its significant downregulation after at least 2 h of heat stress, which was consistent with miR402 induction (Figure 1E and B; Supplementary Figure S1A).
To exclude the possibility that the elevated levels of pri-miR402 and mature miR402 were caused by higher precursor stability after heat stress, A. thaliana seedlings were additionally treated with cordycepin to inhibit transcription and the pri-miR402 half-life was determined. Notably, no significant changes in the stability of the precursors terminated at both proximal and distal polyA sites were observed between stressed and non-stressed plants (Supplementary Figure S2). This shows that the over-accumulation of miR402 in heat stress as compared to the level observed in control plants is not the result of changed pri-miR402 stability.
Moreover, treatments with other selected abiotic stress factors including salinity, mild and severe drought showed the same correlations between the miR402 and pri-miR402 levels, miRNA-carrying intron splicing efficiency and proximal polyadenylation site selection (Supplementary Figure S1C–E).
Active 5΄ splice site and proximal polyA site selection control miR402 accumulation
To investigate the mechanism of miR402 biogenesis regulation, the genomic sequence of At1g77230 was cloned and expressed under the control of the strong 35S promoter in N. benthamiana leaves, since a null mutant of the Arabidopsis At1g77230 gene is not available. First, the effect of miRNA-containing intron 5΄ss inactivation on the miRNA level was determined (Figure 2A). This mutation resulted in the mature miR402 upregulation (Figure 2B and Supplementary Figure S3A). Accordingly to the previously published results (29,30), to exclude the possibility that this observation is caused by the strong 35S promoter, we tested three additional promoter sequences: (i) the native At1g77230 promoter, (ii) the At3g18780 (ACT2) promoter (pACT2) and (iii) the At1g13440 (GAPDH) promoter (pGAPDH). Expression driven by the native At1g77230 promoter resulted in the barely detectable mature miR402 level (Supplementary Figure S3B). However, regardless other promoters used (pACT2, pGAPDH), the mature miR402 level was upregulated upon inactivation of the constitutive 5΄ss of the miRNA-containing intron (Δ5΄ss), as it was observed in the case of the 35S promoter (Figure 2B; Supplementary Figure S3A and B). Therefore, the 35S promoter-driven constructs were used in all subsequent experiments. To rule out the possibility that the oligoU tract that replaced the original 5΄ss sequence of the first intron of the At1g77230 gene may induce some unspecific protein binding, mutations that inactivated only two, three or four nucleotides of the 5΄ss were introduced (Supplementary Figure S3C). Similar to the mutation introducing an oligoU stretch, upregulation of the mature miR402 was observed in all 5΄ss inactivation variants, excluding the effect of the mutation-type engineered (Supplementary Figure S3C).
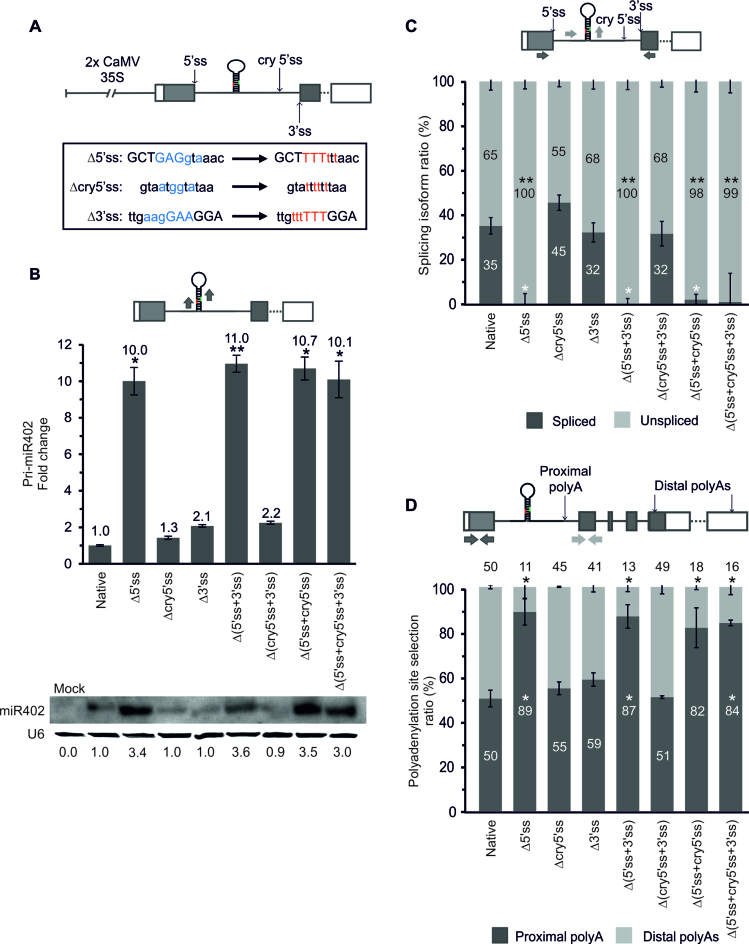
Figure 2.
Active 5΄ss and proximal polyA site selection control miR402 accumulation. (A) The scheme of the At1g77230 gene construct with mutations inactivating 5΄, 3΄ and cryptic 5΄ss depicted in the frame. Blue and red letters indicate nucleotides before and after mutagenesis, respectively. Capital letters indicate exon sequences. (B) The pri-miR402 (upper panel) and mature miR402 (lower panel) levels measured using RT-qPCR and northern blot (U6 used as a loading control), respectively. Numbers below the blot image are relative intensities of the miRNA bands. Mock represents the sample of Nicotiana leaves infiltrated only with buffer (negative control). RT-qPCR analysis of (C) the miR402-bearing intron splicing efficiency and (D) the proximal and distal polyA site usage. Arrows in the upper parts of the panels with the At1g77230 gene scheme depict the primers used (B–D). Δ5΄ss, Δcry5΄ss and Δ3΄ss: mutations inactivating 5΄, cryptic 5΄ and 3΄ splice site, respectively (A–D). Error bars indicate the SD (n = 3), and asterisks indicate a significant difference between the sample and control (*P < 0.05; **P < 0.01; white stars—for spliced or proximal polyA isoforms; black stars—for unspliced and distal polyA isoforms (C-D)).
In the analyzed At1g77230 gene variants with the mutated strong constitutive 5΄ss we observed the activation of a weak cryptic 5΄ss (cry5΄ss) undetected in A. thaliana, and located downstream of the miR402 hairpin. Thus, to test the role of this cry5΄ss and the miRNA-hosting intron 3΄ss in miR402 biogenesis additional constructs were prepared with mutated: (i) the cryptic 5΄ss (Δcry5΄ss), (ii) 3΄ss (Δ3΄ss), (iii) both 5΄ and 3΄ss (Δ(5΄ss+3΄ss)), (iv) Δ(cry5΄ss+3΄ss), (v) Δ(5΄ss+cry5΄ss) and (vi) Δ(5΄ss+cry5΄ss+3΄ss) (Figure 2A). Inactivation of the constitutive 5΄ss in all tested constructs, next to upregulation of mature miR402 level, resulted in the increased level of pri-miR402 (Figure 2B and Supplementary Figure S3A) and caused almost total abolishment of At1g77230 first intron splicing (Figure 2C). Inactivation of the cryptic 5΄ss had no effect on miR402 biogenesis (Figure 2B–D; Supplementary Figure S3A). Additionally, we still noticed the presence of spliced transcripts when the 3΄ss was mutated (Figure 2C) due to the activation of a cryptic 3΄ss located 14 nt downstream from the constitutive 3΄ss (Supplementary Figure S4A). After inactivation of both, constitutive and cryptic 3΄ss (Δ(3΄ss+cry3΄ss)), 23% of the spliced isoform was observed, which was caused by another new cryptic 3΄ss selection (located 9 nt downstream of the first cryptic 3΄ss) (Supplementary Figure S4C). However, the influence of the constitutive 3΄ss and cryptic 3΄ss on miR402 biogenesis was minor in tested conditions (Supplementary Figure S4B).
The analysis of the polyadenylation isoform levels showed that the inhibition of splicing after constitutive 5΄ss inactivation correlated with a significant increase in intronic proximal polyA site selection (Figure 2D and Supplementary Figure S4D), similarly to the polyA site selection shown in the stress experiments in A. thaliana. Moreover, these results were not the effect of higher precursor stability after 5΄ss inactivation, since no significant changes were observed in the half-life of the pri-miR402 ended at both proximal and distal polyA sites and produced from the constructs with active and inactivated 5΄ splice sites (Supplementary Figure S5).
miRNA biogenesis efficiency is dependent on the pre-miRNA location within its host gene
Previously published results showed that the downstream active 5΄ss stimulates the biogenesis of at least three exonic miRNAs: miR161, miR163 and miR172a (29,30). These results are in contrast to our experiments on the intronic miR402, for which the activity of the upstream 5΄ss was found to downregulate the level of this miRNA.
To demonstrate that the position of miRNA stem-loop structure relative to 5΄ss is pivotal for this effect, the miR402 hairpin was moved from the first intron (wild-type At1g77230 gene—Variant A) into the first exon (Variant B) or into the second intron (Variant C) of the miR402 host gene (Figure 3A). In all cases, two versions of each construct were prepared, one with the native first intron 5΄ss (native) and another with the inactivated first intron 5΄ss (Δ5΄ss). All constructs were transiently expressed in tobacco leaves. RT-qPCR analysis revealed the pri-miR402 level upregulation in the case of variants A and B with the inactivated 5΄ss (Figure 3B upper panel). The level of the pri-miR402 located within the second intron did not change when the first intron 5΄ss was mutated (Variant C in Figure 3B upper panel). Moreover, after inactivation of the 5΄ss, the level of mature miR402 increased in Variant A (with the miR402 hairpin present in the first intron) and decreased when the miRNA stem-loop structure was moved to the upstream exon (Variant B) (Figure 3B lower panel). The inactivation of 5΄ss led to first intron splicing inhibition and preferential proximal polyA site selection (Figure 3C and D). We excluded the influence of precursor stability on the miR402 level after 5΄ss inactivation for Variant A (Supplementary Figure S5). Notably, in the case of Variant B, an even-longer half-life of the transcripts polyadenylated at the proximal polyA site was detected when the 5΄ss was mutated; this correlated with the decreased level of miR402 (Figure 3B, lower panel; Supplementary Figure S5A and B). Furthermore, the miR402 originating from the second intron was barely detectable regardless of the presence of functional or inactive first intron 5΄ss, whereas the second intron was always efficiently spliced (Figure 3B, lower panel and E).
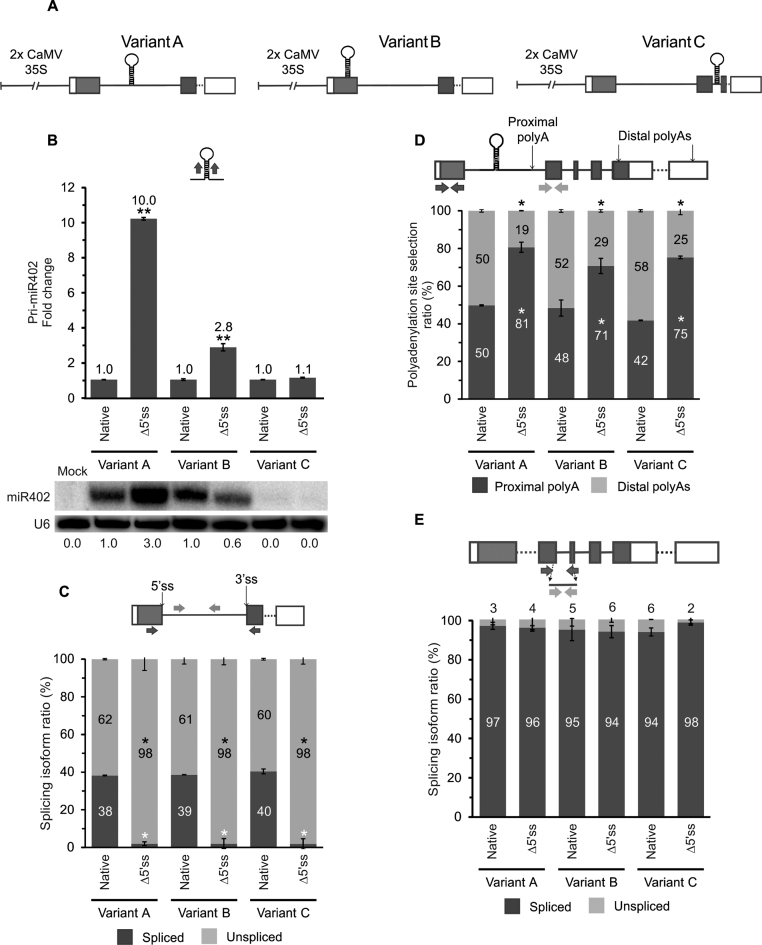
Figure 3.
The efficiency of miRNA biogenesis depends on the pre-miR402 location within its host gene. (A) The scheme of the At1g77230 gene construct variants: A (wild-type), B (the miR402 stem-loop moved into the first exon) and C (the miR402 hairpin moved into the second intron). (B) The pri-miR402 (upper panel) and mature miR402 (lower panel) levels measured using RT-qPCR and northern blot (U6 was used as a loading control), respectively. Numbers below the blot image are relative intensities of the miRNA bands. Mock represents the sample of Nicotiana leaves infiltrated only with buffer (negative control). RT-qPCR analysis of (C) the At1g77230 first intron splicing efficiency, (D) the proximal and distal polyA site selection and (E) At1g77230 second intron splicing efficiency. Arrows in the upper parts of the panels with the At1g77230 gene scheme depict the primers used (B–E). Native and Δ5΄ss mean active and inactivated miR402-carrying intron 5΄ss, respectively. Error bars indicate SD (n = 3), and asterisks indicate a significant difference between the sample and control (*P < 0.05; **P < 0.01; white stars—for spliced or proximal polyA isoforms; black stars—for unspliced and distal polyA isoforms (C-D)).
To conclude, the results presented revealed also that the position of the miRNA stem-loop relative to the proximal 5΄ss determines miRNA biogenesis efficiency. Although the same proximal polyA site was preferentially selected in both A and B variants with an inactivated 5΄ss, this selection did not affect the direction of the miRNA level change. The almost complete abolishment of miR402 production when pre-miR402 was moved to the second intron (Variant C), accompanied by efficient splicing of this intron, suggests that biogenesis from a spliced-out intron is not possible at least in the case of miR402.
SERRATE interacts with U1 snRNP auxiliary proteins
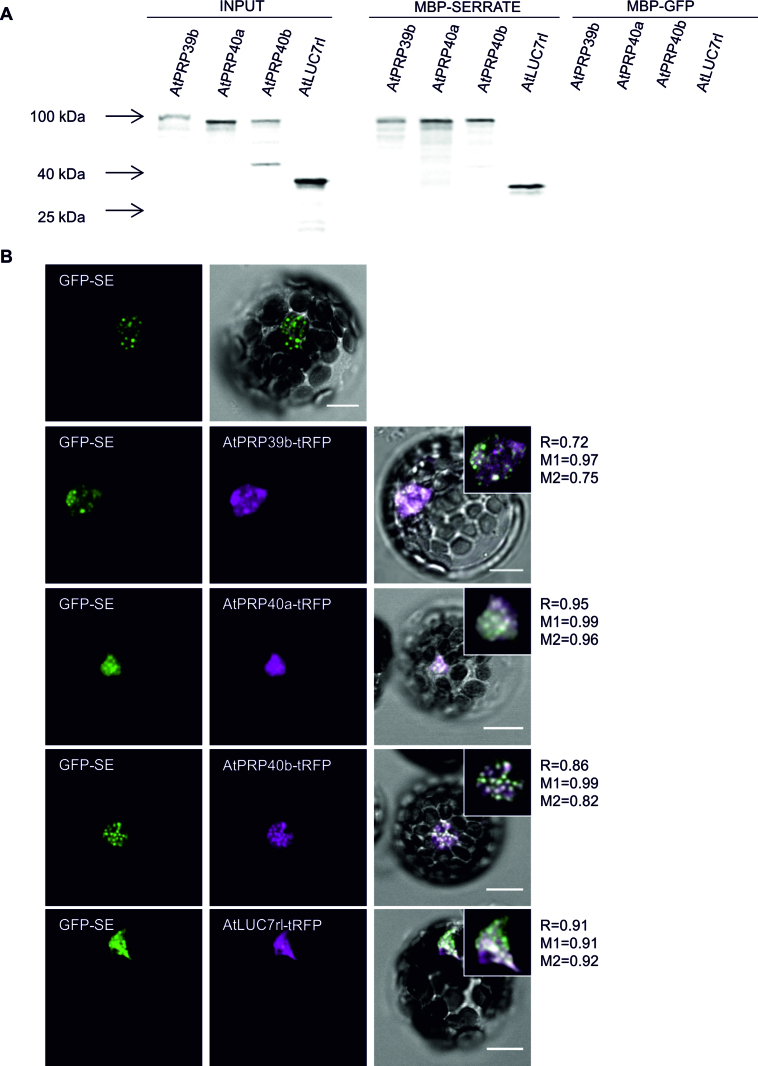
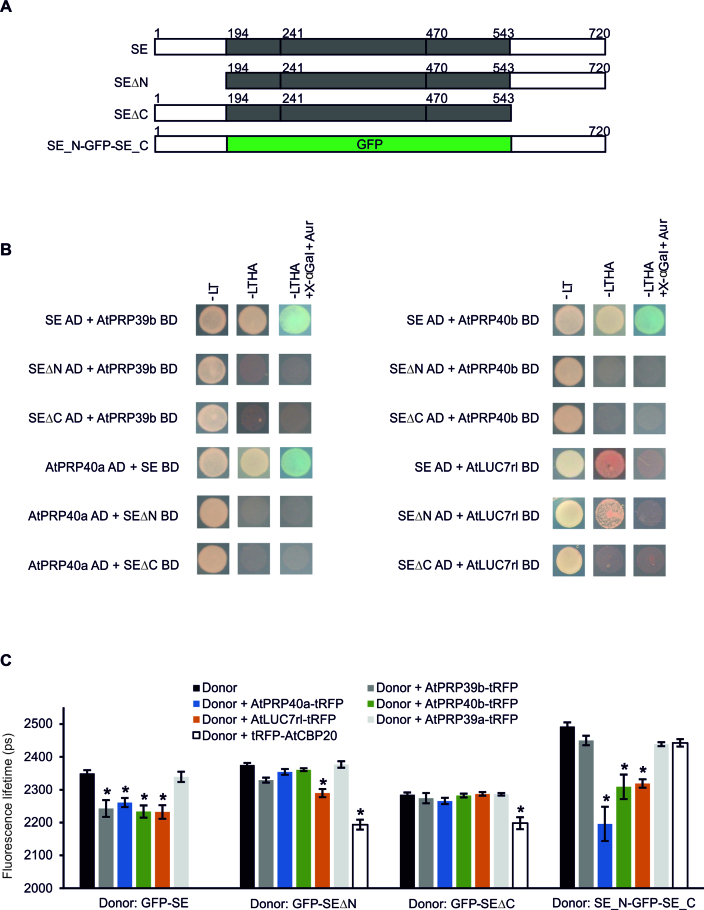
Our results point to the important role of the 5΄ss in miRNA production. Therefore, all known U1 snRNP components were tested for functional connections with SE (At2g27100). A. thaliana orthologs of human and/or yeast U1 snRNP proteins were selected based on experimental data and bioinformatic predictions (56–63). Next, the coding sequences of these proteins were cloned to perform YTH experiments. Among the U1 snRNP components, four auxiliary proteins were found to interact with SE: AtPRP39b (At5g46400), AtPRP40a (At1g44910), AtPRP40b (At3g19670) and AtLUC7rl (At5g51410) (weakly) (Supplementary Figure S6A). Then, the in vitro pull-down assay performed with SE fused to the MBP and [35S]-Met-labeled selected U1 snRNP proteins confirmed the identified interactions (Figure 4A; Supplementary Figure S6B and C). To determine whether SE and its U1 snRNP partners also colocalized in plant cells, SE fused with GFP and one of the interacting U1 snRNP proteins in combination with tagRFP (tRFP) were coexpressed in Arabidopsis protoplasts. We found perfect colocalization of SE and AtPRP39b, AtPRP40a, AtPRP40b and AtLUC7rl (Figure 4B). Next, Förster resonance energy transfer (FRET) analyzed by Fluorescence Lifetime Imaging Microscopy (FLIM) was performed to test if all interactions detected in YTH and in vitro are also taking place in plant protoplasts. FRET-FLIM analysis allows to quantify and visualize direct protein interactions, because FRET can only take place at the nanometer regime. The results confirmed the binding of SE to the four U1 snRNP auxiliary proteins in the plant cell by the observation of donor (GFP-tagged SE) fluorescence lifetime changes in presence of acceptor protein (AtPRP39b-tRFP, AtPRP40a-tRFP, AtPRP40b-tRFP or AtLUC7rl-tRFP) (Figure 5A and C). AtPRP39a-tRFP (which did not interact with SE in YTH) was used as a negative control in these experiments.
Figure 4.
SE interacts with U1 snRNP auxiliary proteins. (A) Analyses of interactions between SE and AtPRP39b, AtPRP40a, AtPRP40b and AtLUC7rl by in vitro pull-down assay. MBP: maltose-binding protein; GFP was used as a negative control; inputs represent one-twentieth of the samples used in the experiment. (B) Colocalization analyses of SE and its U1 snRNP interactors in Arabidopsis thaliana protoplasts. tRFP, tagRFP; R, Pearson's correlation coefficient; M1 and M2, Manders’ overlap coefficients. Insets represent a magnified view of a representative nucleus for each sample. Scale bars = 10 μm.
Figure 5.
Unstructured SE termini are responsible for the interaction with U1 snRNP auxiliary proteins. (A) Schematic structure of the SE variants used in the study. Numbers above the scheme indicate amino acids of the protein (64). (B) YTH analyses of interactions between SE in full-length (SE) or mutated forms (SEΔN and SEΔC) and AtPRP39b, AtPRP40a, AtPRP40b and AtLUC7rl. AD, Gal4 activation domain; BD, Gal4 binding domain; L, Leu; T, Trp; H, His; A, Ade; Aur, Aureobasidin A. (C) FRET-FLIM analyses of protein interactions between SE and its U1 snRNP partners in Arabidopsis thaliana protoplasts. Donor, GFP fused to SE in full length (SE) or mutated forms (SEΔN, SEΔC and SE_N-GFP-SE_C); Acceptor, tagRFP (tRFP) fused to AtPRP39b, AtPRP40a, AtPRP40b, AtLUC7rl, AtPRP39a or AtCBP20; Fluorescence lifetime, lifetime of the donor molecule measured in picoseconds (ps). Error bars indicate the SEM (standard error of the mean, n > 10), and the asterisk indicates a significant difference between the sample in the presence and absence of an acceptor (*P < 0.001).
Two unstructured regions from the N- and C- termini of SERRATE interact with U1 snRNP auxiliary proteins
To determine which part of SE is involved in the interactions with the U1 snRNP proteins, the following constructs were prepared and used in YTH experiments: (i) full-length CDS (SE), and two truncated variants without (ii) N-terminal (SEΔN) or (iii) C-terminal (SEΔC) unstructured tails (Figure 5A) (64). We found that only the AtLUC7rl protein was able to interact weakly with SERRATE lacking the N-terminus (Figure 5B). Both SE terminal fragments were required for interactions with all other U1 snRNP proteins tested. Surprisingly, the confocal microscopy colocalization experiments revealed that the N-truncated variant of SE still colocalized with the U1 snRNP auxiliary proteins studied in the cell nucleus (Supplementary Figures S7A and 8A), whereas the deletion of the SE C-terminal tail resulted in the loss of its colocalization with AtPRP39b-tRFP and AtPRP40b-tRFP (Supplementary Figures S7B). FRET-FLIM analyses confirmed the YTH observations on SE terminal region requirements for direct interaction with U1 snRNP auxiliary proteins in live protoplasts (Figure 5C). AtCBP20 was used as a positive control, since we have previously shown that the core part of SE is sufficient for the CBC binding and we found its perfect colocalization with the SE variants studied (Supplementary Figure S8B) (17). These results additionally show that SE-AtPRP39b and SE-AtPRP40b interactions are necessary for each partner to be targeted into the same nuclear foci.
To exclude the possibility that N- and C-termini must be accompanied by the SE core part to interact with the U1 snRNP partners, SE N- and C-tails were fused to GFP (SE_N-GFP-SE_C) (Figure 5A) and used in FRET-FLIM experiments. The presence of both SE unstructured fragments was sufficient for interactions with AtPRP40a-tRFP, AtPRP40b-tRFP and AtLUC7rl-tRFP (Figure 5C). Although the N- and C-terminal regions of the SE protein were sufficient for the colocalization with all proteins studied in Arabidopsis nuclei (Supplementary Figures S8 and 9), they did not interact directly with AtPRP39b (Figure 5C).
The interactions between SE and U1 snRNP auxiliary proteins affect miRNA biogenesis
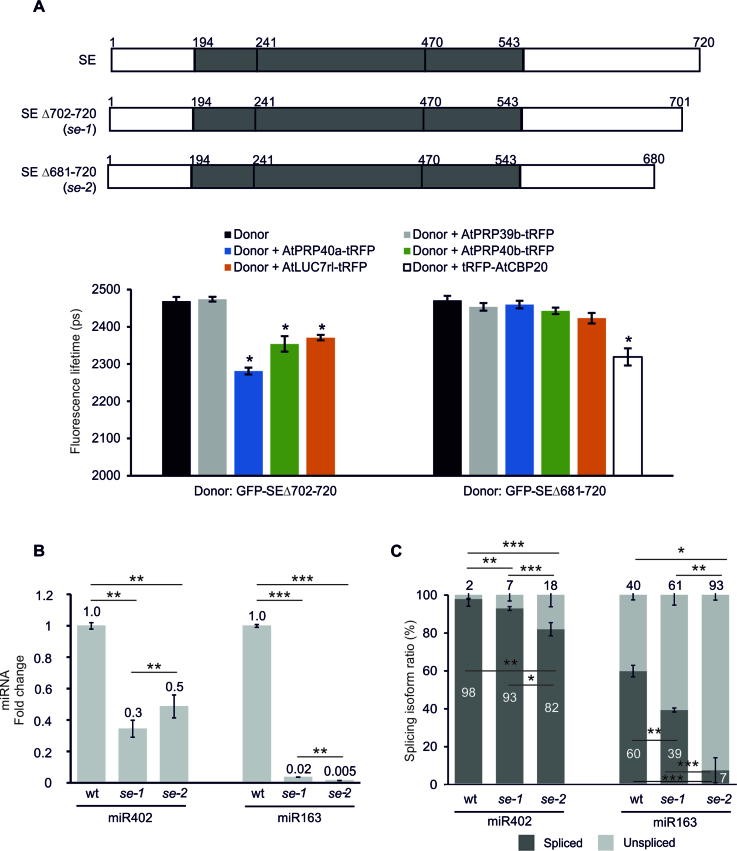
The experiments presented thus far suggest that SE-U1 snRNP auxiliary protein interactions may stimulate microprocessor activity when the miRNA stem-loop structure is located between the 5΄ end of the pri-miRNA and the 5΄ss but inhibit its action when the miRNA hairpin is located downstream of the 5΄ss. To support these assumptions, we used two viable hypomorphic mutants, se-1 and se-2, that lack 20 and 40 amino acid residues, respectively, from the C-terminus (44,45). Although the levels of many miRNAs are changed in both these mutants, the SE C-terminus is not required for binding core microprocessor components (DCL1 and HYL1) (36,45,64–66). Thus, FRET-FLIM analyses were performed to determine whether these truncated SE proteins still form a complex with the U1 snRNP auxiliary proteins identified as SE partners. The SE version corresponding to se-1 colocalized and interacted with AtPRP40a, AtPRP40b and AtLUC7rl, but the variant present in the se-2 mutant plants was no longer able to bind to any of the tested U1 snRNP auxiliary proteins (Figure 6A and Supplementary Figure S10). Thus, in the se-2 plants, there is no communication between SE and its U1 snRNP partners, however this SE variant still interacts with AtCBC (Figure 6A).
Figure 6.
SE is a key protein connecting the microprocessor and the spliceosome in the plant cell. (A, upper panel) Schematic structure of the SE variants used in the study, corresponding to the full length protein (SE) and its variants in se-1 and se-2 mutant plants (44,45). Numbers above the scheme indicate the amino acids of the protein (64). (A, lower panel) FRET-FLIM analyses of interactions between SE variants and U1 snRNP proteins in Arabidopsis thaliana protoplasts. Donor, GFP fused to full-length SE (SE) or to the mutated forms (SEΔ702-720 and SEΔ681-720); Acceptor, tagRFP (tRFP) fused to AtPRP39b, AtPRP40a, AtPRP40b, AtLUC7rl or AtCBP20; Fluorescence lifetime, lifetime of the donor molecule measured in picoseconds (ps). Error bars indicate the SEM (n > 10), and the asterisk indicates a significant difference between the sample in the presence and absence of an acceptor (*P < 0.001). RT-qPCR analysis of (B) the mature miR402 and miR163 levels and (C) splicing efficiency of the miR402-hosting intron and the MIR163 intron, respectively, in the se-1 and se-2 mutants and wild-type plants. (B and C) Error bars indicate SD (n = 3), and asterisks indicate a significant difference between the samples and control (*P < 0.05; **P < 0.01; ***P < 0.001).
Next, the levels of intronic miR402 and exonic miR163 (At1g66725) (located downstream and upstream of the 5΄ss, respectively) were compared in both se mutants to test the effect of the presence (se-1) and the loss (se-2) of SE-U1 snRNP communication on the miRNA biogenesis. As expected, the level of mature miR402 was downregulated in se-1 compared to the wt plants due to the less-efficient miRNA biogenesis (44) (Figure 6B). Although in se-2 (similar to the se-1 mutant), the microprocessor activity is also impaired (45), a higher level of miR402 was observed when compared to se-1 plants (Figure 6B). This was correlated with the stronger splicing inhibition of the miRNA-carrying intron in se-2 as compared to the se-1 (Figure 6C). These results were accompanied by the upregulated and unchanged pri-miR402 levels in the se-1 (36) and se-2 (Supplementary Figure S11A) mutants, respectively. Notably, the loss of SE-U1 snRNP auxiliary protein communication had the opposite effect on miR163 production. In se-2 mutant plants, the splicing of the pre-miR163-downstream intron was the most inhibited and the lowest level of mature miRNA was detected (Figure 6B and C). The level of pri-miR163 was not affected in either se-1 (36) or se-2 mutants as compared to wild-type plants (Supplementary Figure S11A). This data indicated that the SE-U1 snRNP interactions are responsible for the regulation of splicing-related miRNA biogenesis. The lack of interactions between SE and U1 snRNP auxiliary proteins in the se-2 mutant resulted in the most-severely inhibited splicing of the miR402-carrying intron, thus allowing more mature miR402 to accumulate. In contrast, the biogenesis of exonic miR163 was the most impaired in the se-2 mutant; this was correlated with the decreased level of splicing of the pre-miRNA-downstream intron (Figure 6). Moreover, the maturation of two other miRNAs (intronic miR1888a that is located within the first intron of At5g21100, and exonic miR171b which is encoded by an intron-containing MIR171b (At1g11735)) is regulated by the similar mechanism. The higher splicing inhibition of the miRNA-carrying intron in se-2 plants was accompanied by miR1888a level upregulation as compared to the level detected in the se-1 mutant (Supplementary Figure S11B). For the exonic miR171b, we also observed splicing inhibition in se-2 plants, which was correlated with even stronger miR171b level downregulation than in the se-1 mutant (Supplementary Figure S11C). As in the case of miR163 and miR402, we did not observe any changes in the pri-miR1888a and pri-miR171b levels in se-2 (Supplementary Figure S11B and C); however, both analyzed pri-miRNAs were upregulated in the se-1 mutant (36).
Thus, to conclude, the deleted part of the SE protein in the se-2 mutant is mostly responsible for the interaction of SE with U1 snRNP partners and its presence stimulates exonic miRNA production and inhibits intronic miRNA biogenesis (at least in the cases of the miRNAs tested).
DISCUSSION
5΄ splice site affects the biogenesis of miRNAs encoded by intron-containing genes
The data presented in this study and in our previous paper demonstrate that the active 5΄ss influences the biogenesis of plant miRNAs derived from intron-containing genes (29). The 5΄ss located downstream of the miRNA stem-loop structure stimulates mature miRNA production. In contrast, for at least two studied intronic miRNAs (miR402 and miR1888a), efficient splicing of the miRNA-hosting intron had an inhibitory effect on mature miRNA production. However, when the intronic pre-miR402 was moved to the upstream exon, the active 5΄ss enhanced miRNA maturation, as has been shown for the biogenesis of exonic miR163, miR161 and miR172a (29,30). Thus, the stimulatory or inhibitory effect of the 5΄ss on miRNA accumulation clearly depends on the reciprocal spatial relation between the positions of the miRNA stem-loop structure and the nearest active 5΄ss and is not influenced by the precursor stability.
The influence of splicing on miRNA production described here can explain the changes in intronic miR402 levels upon various stress treatments. The level of mature miR402 was elevated primarily under the analyzed stress conditions compared to the control. This increase correlated with the lower splicing efficiency of the miRNA-carrying intron and might be caused by stress-induced modulation of protein–protein and RNA–protein interaction stability and/or secondary and tertiary RNA structure changes (67,68). Additionally, the transcription rate might play a role in the regulation of splicing, polyadenylation and (ultimately) the level of miRNA precursor. Indeed, the influence of transcription elongation rate on splicing and vice versa has been reported (69–71). Moreover, a broad transcriptional response of pri-miRNAs has already been detected for common climate change-related stresses, like drought, heat and salinity (9). Interestingly, no changes in At1g77230 fourth intron splicing efficiency were observed in any of the stresses studied. Therefore, a mechanism exists that differently affects At1g77230 intron splicing under stress conditions.
Notably, not all intronic miRNAs are upregulated under stress treatment. For example, the level of miR400 decreased under various conditions, that seems to correlate with the splicing inhibition of an miRNA-carrying intron (9,38). Thus, we can conclude, that the interplay between splicing and intronic miRNA production can be also affected differently by additional factors of transcriptional and/or post-transcriptional regulation. However, the splicing efficiency clearly has a decisive role in the production of either functional miR402 or mRNA of its host gene. Therefore, this mechanism is significant for gene expression regulation.
In addition to the miR402-hosting intron splicing inhibition observed under the tested conditions, preferential selection of the intronic, proximal polyA site was also detected. This effect is likely caused by the lack of efficient U1 snRNP binding to the 5΄ss and the consequent abolishment of the U1 snRNP role in protecting pre-mRNAs from premature cleavage and polyadenylation, as shown previously in HeLa cells (72). Our transient expression experiments in tobacco confirmed the influence of the 5΄ss activity on polyA site selection. These results show that 5΄ss inactivation stimulates the use of the proximal polyadenylation site but does not always result in upregulation of miR402. When the miR402 hairpin was located within the first intron, enhanced mature miRNA production was observed after 5΄ss mutation, whereas the relocation of the miR402 stem-loop to the upstream exon resulted in miR402 downregulation, despite efficient selection of the same proximal polyA site in both cases. This observation confirmed the data concerning exonic miR163 production, which showed that after 5΄ss inactivation, the proximal intronic polyA site was preferentially selected, resulting in miRNA downregulation (29). In both cases, intronic miR402 and exonic miR163 splicing inhibition led to the selection of alternative proximal polyA sites and the generation of independent, shorter, intron-less transcripts. These results indicated that the 5΄ss and splicing activity, not polyadenylation site selection, primarily determine miRNA biogenesis efficiency.
Interestingly, the pri-miR402 abundance increases in both Variant A (the miR402 hairpin is located in the intron) and B (the miR402 hairpin is located in the upstream exon) when the 5΄ss is inactivated. However, the level of mature miR402 from Variant B but not A decreases upon inactivation of the 5΄ss. Since the transcript derived from Variant A with the active 5΄ss is efficiently spliced, the amount of pri-miRNA measured by the amount of the unspliced transcript is very low. We postulate that this is the reason for the high upregulation of the miR402 primary precursor level in Variant A when the 5΄ss is inactivated and splicing totally inhibited. In Variant B, however, the miR402 biogenesis is less efficient and less miR402 is accumulated after 5΄ss inactivation. The upregulated level of pri-miR402 in this case is due to the less-efficient biogenesis of miR402 from variant B transcripts. The upregulated pri-miRNA levels caused by the impaired microprocessor activity have already been reported for many miRNA biogenesis mutants; i.e. se-1, hyl1-2, dcl1-7 and cbc (36). Thus, the increased pri-miR402 level in Variant B after 5΄ss mutation represents an unprocessed precursor.
The crosstalk between CBC/microprocessor/U1 snRNP plays a crucial role in regulating miRNA biogenesis
AtCBP20, AtCBP80 and SE are involved in both splicing and miRNA biogenesis (15–17,73–74). However, the mechanism of this dual function of SE and AtCBC and the identities of their interacting partners in the splicing machinery are unknown. Previously published data showed that SE forms a complex with AtCBC and interacts with DCL1, HYL1 and the majority of the recently identified players in miRNA biogenesis (17,19,20,22,23,25,75). Thus, SE is a good candidate for a key scaffold protein involved in plant microprocessor and spliceosome communication. The presented experiments provide clear evidence for the direct interactions between SE and four auxiliary U1 snRNP proteins; i.e. AtPRP39b, AtPRP40a, AtPRP40b and AtLUC7rl in vitro and in the plant cell. Interestingly, human and yeast orthologs of SE-identified spliceosome partners have been shown to interact with other RNA metabolism machineries, including BP-binding proteins (Msl5p, BBP, SF1), the cleavage factor Im complex (CPSF6), the nuclear exosome targeting complex (ZCCHC8) and the exon junction complex (RNPS1) (76–80). In addition, PRP40 interacts with the CTD domain of RNA polymerase II (63,81,82). Thus, we suggest that AtPRP39b, AtPRP40a, AtPRP40b and AtLUC7rl form a platform for protein-protein interactions and, together with SE, are responsible for the crosstalk between the plant spliceosome and the microprocessor.
In 2011, SE was shown to consist of the structured core (interacting with HYL1) and two unstructured N- and C-terminal fragments (64). The core part of SE interacts also with AtCBC (17). Furthermore, direct binding of SE to DCL1 requires its core fragment and the N-terminal unstructured part that is also responsible for RNA binding (66). The presented results indicate that both SE N- and C- unstructured regions are involved in the interactions with U1 snRNP auxiliary proteins and function as an anchor for their binding. However, these regions were not sufficient for AtPRP39b contact, which suggests that the core part of SE and/or other U1 snRNP components must be additionally present to enable the binding of this protein. We presume that four SE partners of U1 snRNP, and their different requirements for these interactions stabilize direct microprocessor-spliceosome communication. These findings may also reflect the special localization of these auxiliary proteins within the U1 snRNP. Alternatively, our observations may indicate the diversity of the U1 snRNP composition itself (the high similarity of AtPRP40a and AtPRP40b proteins indicate that they could replace each other within this complex) and in complexes with other machineries depending on the RNA substrate and/or plant growth conditions.
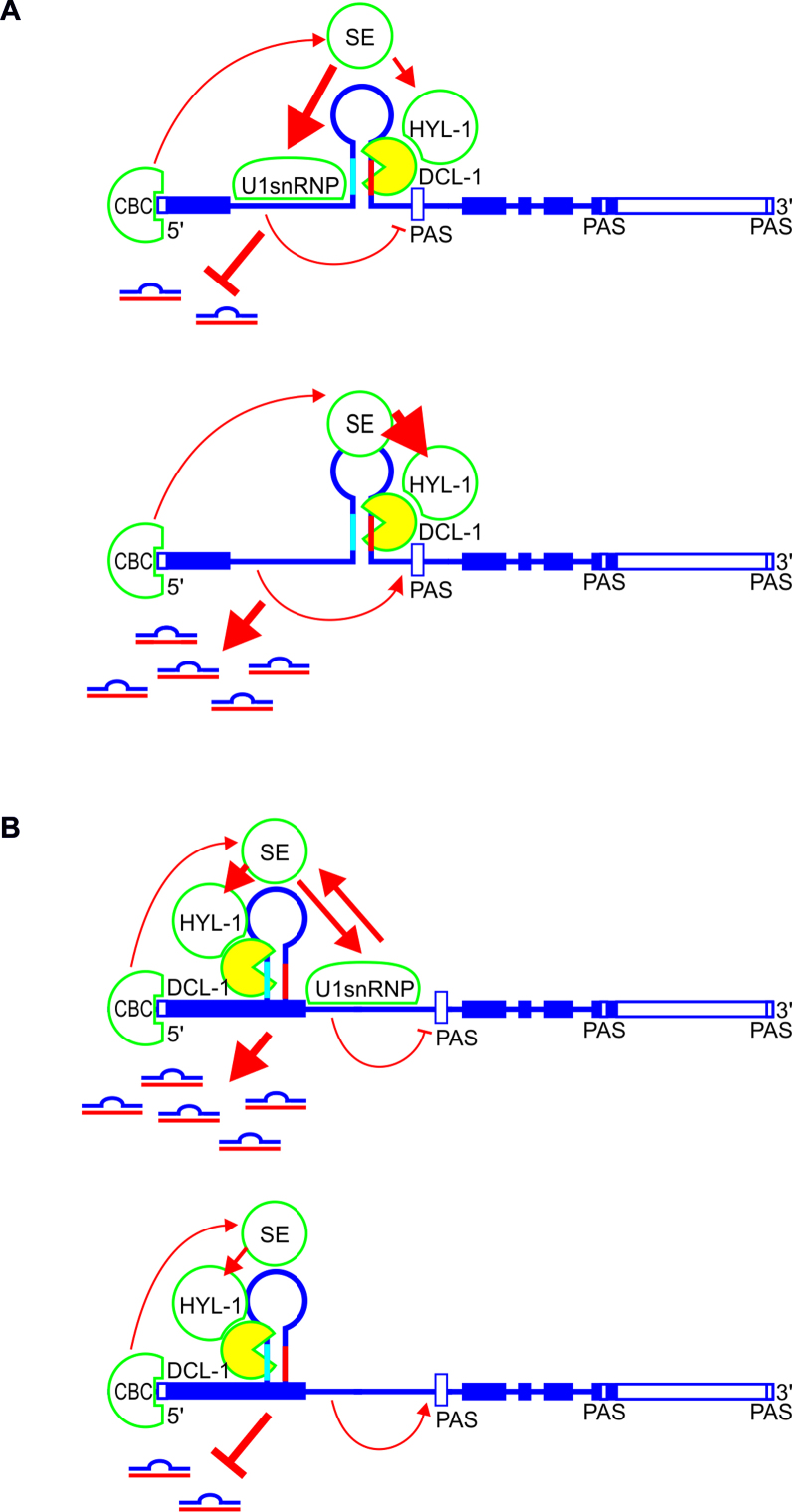
Based on our findings, we propose that the direct interactions among the microprocessor, the 5΄ss and the spliceosome influence selected intronic and exonic miRNA biogenesis (Figure 7). CBC binds to the cap of a primary transcript containing a miRNA stem-loop structure within the first intron. Next, SE interacts with CBC and U1 snRNP through the structured core and unstructured fragment, respectively. As a result, SE has rather limited access to the microprocessor complex located downstream of the 5΄ss, enhancing splicing efficiency and impairing intronic miR402 and miR1888a production. Consequently, 5΄ss inactivation inhibits U1 snRNP binding, allows SE to fully participate in microprocessor formation and pre-miRNA binding, and leads to efficient miRNA production (Figure 7A). In the case of exonic miR163 and miR171b, immediately after CBC recruitment to the cap, the miRNA stem-loop structure is formed and microprocessor proteins, including SE, bind to the hairpin. Next, U1 snRNP binding to the active 5΄ss stabilizes SE interactions with other proteins involved in miRNA biogenesis and enhances mature miRNA production (Figure 7B). This model is strongly supported by the stimulatory and inhibitory effects of SE - U1 snRNP interaction impairment during analyzed intronic and exonic miRNA biogenesis, respectively, in the se-2 mutant plants (Figure 6 and Supplementary Figure S11). Thus, SE is a crucial protein in spliceosome and plant microprocessor communication. Moreover, SE-U1 snRNP interplay modulates the processing of analyzed intron-containing pri-miRNAs, and its stimulatory or inhibitory effect depends on the position of the miRNA stem-loop structure relative to the closest active 5΄ss.
Figure 7.
A model of the interplay between the microprocessor, 5΄ss and U1 snRNP during biogenesis of selected intronic (A) and exonic (B) miRNA in plants. Boxes represent exons, and lines depict introns. Light blue and red parts of the hairpin mark miRNA and miRNA*, respectively. Thick arrows show strong connections, while thin arrows show weak connections. The no-headed arrow points to the inhibitory effect. Open boxes mark proximal and distal polyA sites (PAS). CBC: CAP-BINDING COMPLEX; SE: SERRATE; HYL1: HYPONASTIC LEAVES 1; DCL1: DICER-LIKE1.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Prof. Gordon G. Simpson for help in polyadenylation site analyses, to Dr Agnieszka Ludwikow for providing yeast strains, plasmids and help in the YTH analyses and to Prof. Aleksandra Obrepalska-Steplowska for supplying N. benthamiana plants and seeds.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Centre [2012/04/M/NZ2/00127 to Z.S.K., 2013/10/A/NZ1/00557 to A.J., 2013/11/B/NZ1/02099 to Z.S.K., 2012/05/N/NZ2/00880 to A.S., 2012/05/N/NZ2/00955 to K.K., 2015/16/T/NZ1/00022 to K.K., 2015/16/T/NZ1/00026 to A.S.]; Polish Ministry of Science and Higher Education [01/ KNOW2/2014 to KNOW RNA Research Centre in Poznan]; Faculty of Biology at Adam Mickiewicz University (Badania Statutowe). Funding for open access charge: Polish Ministry of Science and Higher Education [01/ KNOW2/2014].
Conflict of interest statement. None declared.
REFERENCES
- 1.Carrington J.C., Ambros V.. Role of microRNAs in plant and animal development. Science. 2003; 301:336–338. [DOI] [PubMed] [Google Scholar]
- 2.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009; 136:669–687. [DOI] [PubMed] [Google Scholar]
- 3.Reis R.S., Hart-Smith G., Eamens A.L., Wilkins M.R., Waterhouse P.M.. Gene regulation by translational inhibition is determined by Dicer partnering proteins. Nat. Plants. 2015; 1:14027. [DOI] [PubMed] [Google Scholar]
- 4.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004; 303:2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallory A.C., Dugas D.V., Bartel D.P., Bartel B.. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 2004; 14:1035–1046. [DOI] [PubMed] [Google Scholar]
- 6.Mallory A.C., Bartel D.P., Bartel B.. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005; 17:1360–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sunkar R., Zhu J.K.. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004; 16:2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kruszka K., Pieczynski M., Windels D., Bielewicz D., Jarmolowski A., Szweykowska-Kulinska Z., Vazquez F.. Role of microRNAs and other sRNAs of plants in their changing environments. J. Plant Physiol. 2012; 169:1664–1672. [DOI] [PubMed] [Google Scholar]
- 9.Barciszewska-Pacak M., Milanowska K., Knop K., Bielewicz D., Nuc P., Plewka P., Pacak A., Vasquez F., Karlowski W., Jarmolowski A. et al. . Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front. Plant Sci. 2015; 6:410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B. MicroRNA: a new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 2015; 66:1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park M.Y., Wu G., Gonzalez-Sulser A., Vaucheret H., Poethig R.S.. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:3691–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C.. Expression of Arabidopsis miRNA genes. Plant Physiol. 2005; 138:2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P.. MicroRNAs in plants. Genes Dev. 2002; 16:1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong Z., Han M.H., Federoff N.. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:9970–9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laubinger S., Sachsenberg T., Zeller G., Busch W., Lohmann J.U., Ratsch G., Weigel D.. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:8795–8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raczynska K.D., Simpson C.G., Ciesiolka A., Szewc L., Lewandowska D., McNicol J., Szweykowska-Kulinska Z., Brown J.W., Jarmolowski A.. Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2010; 38:265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raczynska K.D., Stepien A., Kierzkowski D., Kalak M., Bajczyk M., McNicol J., Simpson C.G., Szweykowska-Kulinska Z., Brown J.W., Jarmolowski A.. The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2014; 42:1224–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaabane S.B., Liu R., Chinnusmay V., Kwon Y., Park J.H., Kim S.Y., Zhu J.K., Yang S.W., Lee B.H.. STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res. 2012; 41:1984–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manavella P.A., Hagmann J., Ott F., Laubinger S., Franz M., Macek B., Weigel D.. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell. 2012; 151:859–870. [DOI] [PubMed] [Google Scholar]
- 20.Ren G., Xie M., Dou Y., Zhang S., Zhang Ch., Yu B.. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:12817–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhan X., Wang B., Li H., Liu R., Kalia R.K., Zhu J.K., Chinnusamy V.. Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:18198–18203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L., Song X., Gu L., Li X., Cao S., Chu C., Cui X., Chen X., Cao X.. NOT2 proteins promote polymerase II-dependent transcription and interact with multiple microRNA biogenesis factors in Arabidopsis. Plant Cell. 2013; 25:715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speth C., Willing E.M., Rausch S., Schneeberger K., Laubinger S.. RACK1 scaffold proteins influence miRNA abundance in Arabidopsis. Plant J. 2013; 6:433–445. [DOI] [PubMed] [Google Scholar]
- 24.Wu X., Shi Y., Li J., Xu L., Fang Y., Li X., Qi Y.. A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Res. 2013; 23:645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S., Liu Y., Yu B.. PRL1, an RNA-binding protein, positively regulates the accumulation of miRNAs and siRNAs in Arabidopsis. PLOS Genet. 2014; 10:e1004841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B., Duan Ch.G, Wang X., Hou Y.J., Yan J., Gao C., Kim J.H., Zhang H., Zhu J.K.. HOS1 regulates Argonaute1 by promoting transcription of the microRNA gene MIR168b in Arabidopsis. Plant J. 2015; 81:861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koster T., Meyer K., Weinholdt C., Smith L.M., Lummer M., Speth C., Grosse I., Weigel D., Staiger D.. Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic Acids Res. 2014; 42:9925–9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen T., Cui P., Xiong L.. The RNA-binding protein HOS5 and serine/arginine-rich proteins RS40 and RS41 participate in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 2015; 43:8283–8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bielewicz D., Kalak M., Kalyna M., Windels D., Barta A., Vazquez F., Szweykowska-Kulinska Z., Jarmolowski A.. Introns of plant pri-miRNAs enhance miRNA biogenesis. EMBO Rep. 2013; 14:622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwab R., Speth C., Laubinger S., Voinnet O.. Enhanced microRNA accumulation through stem loop-adjacent introns. EMBO Rep. 2013; 14:615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruszka K., Pacak A., Swida-Barteczka A., Stefaniak A.K., Kaja E., Sierocka I., Karlowski W., Jarmolowski A., Szweykowska-Kulinska Z.. Developmentally regulated expression and complex processing of barley pri-microRNAs. BMC Genomics. 2013; 14:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kruszka K., Pacak A., Swida-Barteczka A., Nuc P., Alaba S., Wroblewska Z., Karlowski W., Jarmolowski A., Szweykowska-Kulinska Z.. Transcriptionally and post-transcriptionally regulated microRNAs in heat stress response in barley. J. Exp. Bot. 2015; 65:6123–6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szweykowska-Kulinska Z., Jarmolowski A., Vazquez F.. The crosstalk between plant microRNA biogenesis factors and the spliceosome. Plant Signal. Behav. 2013; 8:e26955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurihara Y., Watanabe Y.. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szarzynska B., Sobkowiak L., Pant B.D., Balazadeh S., Scheible W.R., Mueller-Roeber B., Jarmolowski A., Szweykowska-Kulinska Z.. Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res. 2009; 7:3083–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zielezinski A., Dolata J., Alaba S., Kruszka K., Pacak A., Swida-Barteczka A., Knop K., Stepien A., Bielewicz D., Pietrykowska H. et al. . mirEX 2.0 - an integrated environment for expression profiling of plant microRNAs. BMC Plant Biol. 2015; 15:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown J.W., Marshall D.F., Echeverria M.. Intronic noncoding RNAs and splicing. Trends Plant Sci. 2008; 13:335–342. [DOI] [PubMed] [Google Scholar]
- 38.Yan K., Liu P., Wu C.A., Yang G.D., Xu R., Guo Q.H., Huang J.G., Zheng C.C.. Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell. 2012; 48:521–531. [DOI] [PubMed] [Google Scholar]
- 39.Janas M.M., Khaled M., Schubert S., Bernstein J.G., Golan D., Veguilla R.A., Fisher D.E., Shomron N., Levy C., Novina C.D.. Feed-forward microprocesing and splicing activities at a microRNA-containing intron. PLoS Genet. 2011; 7:e1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramalingam P., Palanichamy J.K., Singh A., Das P., Bhagat M., Kassab M.A., Sinha S., Chattopadhyay P.. Biogenesis of intronic miRNAs located in clusters by independent transcription and alternative splicing. RNA. 2014; 20:76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agranat-Tamir L., Shomron N., Sperling J., Sperling R.. Interplay between pre-mRNA splicing and microRNA biogenesis within the supraspliceosome. Nucleic Acids Res. 2014; 42:4640–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng Y., Shao Ch.. Large-scale identification of mirtrons in Arabidopsis and rice. PLoS One. 2012; 7:e31163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joshi P.K., Gupta D., Nandal U.K., Khan Y., Mukherjee S.K., Sanan-Mishra N.. Identification of mirtrons in rice using MirtronPred: a tool for predicting plant mirtrons. Genomics. 2012; 99:370–375. [DOI] [PubMed] [Google Scholar]
- 44.Prigge M.J., Wagner D.R.. The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell. 2001; 13:1263–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grigg S.P., Canales C., Hay A., Tsiantis M.. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature. 2005; 437:1022–1026. [DOI] [PubMed] [Google Scholar]
- 46.Seeley K.A., Byrne D.H., Colbert J.T.. Red light-independent instability of oat phytochrome mRNA in vivo. Plant Cell. 1992; 4:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Curtis M.D., Grossniklaus U.A.. Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003; 133:462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bielewicz D., Dolata J., Zielezinski A., Alaba S., Szarzynska B., Szczesniak M., Jarmolowski A., Szweykowska-Kulinska Z., Karlowski W.M.. mirEX: a platform for comparative exploration of plant pri-miRNA expression data. Nucleic Acids Res. 2011; 40:D191–D197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu F.H., Shen S.C.H., Lee L.Y., Lee S.H., Chan M.T., Lin C.H.S. Tape-Arabidopsis Sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods. 2009; 5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bucherl C., Aker J., de Vries S., Borst JW.. Probing protein-protein Interactions with FRET-FLIM. Methods Mol. Biol. 2010; 655:389–399. [DOI] [PubMed] [Google Scholar]
- 51.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. . Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bucherl C.H.A., van Esse G.W., Kruis A., Luchtenberg J., Westphal A.H., Aker J., van Hoek A., Albrecht C., Borst JW., de Vires S.C.. Visualization of BRI1 and BAK1(SERK3) membrane receptor heterooligomers during brassinosteroid signaling. Plant Physiol. 2013; 162:1911–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baev V., Milev I., Naydenov M., Vachev T., Apostolova E., Mehterov N., Gozmanva M., Minkov G., Sablok G., Yahubyan G.. Insight into small RNA abundance and expression in high- and low-temperature stress response using deep sequencing in Arabidopsis. Plant Physiol. Biochem. 2014; 84:105–114. [DOI] [PubMed] [Google Scholar]
- 54.Sherstnev A., Duc C., Cole C., Zacharaki V., Hornyik C., Ozsolak F., Milos P.M., Barton J.G., Simpson G.G.. Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat. Struct. Mol. Biol. 2012; 19:845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim J.Y., Kwak K.J., Jung H.J., Lee H.J., Kang H.. MicroRNA402 affects seed germination of Arabidopsis thaliana under stress conditions via targeting DEMETER-LIKE Protein 3 mRNA. Plant Cell Physiol. 2010; 51:1079–1083. [DOI] [PubMed] [Google Scholar]
- 56.Simpson G.G., Clark G.P., Rothnie H.M., Boelens W., van Venrooij W., Brown J.W.. Molecular characterization of the spliceosomal proteins U1A and U2B' from higher plants. EMBO J. 1995; 14:4540–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Golovkin M., Reddy A.S.. Structure and expression of a plant U1 snRNP 70K gene: alternative splicing of U1 snRNP 70K pre-mRNAs produces two different transcripts. Plant Cell. 1996; 8:1421–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neubauer G., Gottschalk A., Fabrizio P., Seraphin B., Luehrmann R., Mann M.. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krecic A.M., Swanson M.S.. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 1999; 11:363–371. [DOI] [PubMed] [Google Scholar]
- 60.Lorkovic Z.J., Hilscher J., Barta A.. Use of fluorescent protein tags to study nuclear organization of the spliceosomal machinery in transiently transformed living plant cells. Mol. Biol. Cell. 2004; 15:3233–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang B.B., Brendel V.. The ASRG database: identification and survey of Arabidopsis thaliana genes related to pre-mRNA splicing. Genome Biol. 2004; 5:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang C., Tian Q., Hou Z., Mucha M., Aukerman M., Olsen O.A.. The Arabidopsis thaliana AT PRP39-1 gene, encoding a tetratricopeptide repeat protein with similarity to the yeast pre-mRNA processing protein PRP39, affects flowering time. Plant Cell Rep. 2007; 26:1357–1366. [DOI] [PubMed] [Google Scholar]
- 63.Kang C.H.H., Fenga Y., Vikrama M., Jeongb I.S., Leeb J.R., Bahk J.D., Yun D.J., Lee S.Y., Koiwa H.. Arabidopsis thaliana PRP40s are RNA polymerase II C-terminal domain-associating proteins. Arch. Biochem. Biophys. 2009; 484:30–38. [DOI] [PubMed] [Google Scholar]
- 64.Machida S., Chen H.J., Yuan Y.A.. Molecular insights into miRNA processing by Arabidopsis thaliana SERRATE. Nucleic Acids Res. 2011; 39:7828–7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lobbes D., Rallapalli D., Schmidt D.D., Martin C., Clarke J.. SERRATE: a new player on the plant microRNA scene. EMBO Rep. 2006; 7:1052–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iwata Y., Takahashi M., Fedoroff N.V., Hamdan S.M.. Dissecting the interactions of SERRATE with RNA and DICER-LIKE 1 in Arabidopsis microRNA precursor processing. Nucleic Acids Res. 2013; 41:9129–9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chursov A., Kopetzky S.J., Leshchiner I., Kondofersky I., Theis F.J., Frishman D., Shneider A.. Specific temperature-induced perturbations of secondary mRNA structures are associated with the cold-adapted temperature-sensitive phenotype of influenza A virus. RNA Biol. 2012; 9:1266–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feig A.L. Studying RNA-RNA and RNA-protein interactions by isothermal titration calorimetry. Methods Enzymol. 2009; 468:409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwek K.Y., Murphy S., Furger A., Thomas B., O'Gorman W., Kimura H., Proudfoot N.J., Akoulitchev A.. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat. Struct. Mol. Biol. 2002; 9:800–805. [DOI] [PubMed] [Google Scholar]
- 70.de la Mata M., Alonso C.R., Kadener S., Fededa J.P., Blaustein M., Pelisch F., Cramer P., Bentley D., Kornblihtt A.R.. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003; 12:525–532. [DOI] [PubMed] [Google Scholar]
- 71.Dolata J., Guo Y., Kolowerzo A., Smolinski D., Brzyzek G., Jarmolowski A., Swiezewski Sz.. NTR1 is required for transcription elongation checkpoints at alternative exons in Arabidopsis. EMBO J. 2015; 34:544–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaida D., Berg M.G., Younis I., Kasim M., Singh L.N., Wan L., Dreyfuss G.. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010; 468:664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim S., Yang J.Y., Xu J., Jang I.C., Prigge M.J., Chua N.H.. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary MicroRNAs. Plant Cell Physiol. 2008; 49:1634–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kierzkowski D., Kmieciak M., Piontek P., Wojtaszek P., Szweykowska-Kulinska Z., Jarmolowski A.. The Arabidopsis CBP20 targets the cap-binding complex to the nucleus, and is stabilized by CBP80. Plant J. 2009; 59:814–825. [DOI] [PubMed] [Google Scholar]
- 75.Fang Y., Spector D.L.. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 2007; 17:818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abovich N., Rosbash M.. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997; 89:403–412. [DOI] [PubMed] [Google Scholar]
- 77.Colland F., Jacq X., Trouplin V., Mougin C., Groizeleau C., Hamburger A., Meil A., Wojcik J., Legrain P., Gauthier J.M.. Functional proteomics mapping of a human signaling pathway. Genome Res. 2004; 14:1324–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sakashita E., Tatsumi S., Werner D., Endo H., Mayeda A.. Human RNPS1 and its associated factors: a versatile alternative pre-mRNA splicing regulator in vivo. Mol. Cell. Biol. 2004; 24:1174–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ingham R.J., Colwill K., Howard C., Dettwiler S., Lim C.S.H., Yu J., Hersi K., Raaijmakers J., Gish G., Mbamalu G. et al. . WW domains provide a platform for the assembly of multiprotein networks. Mol. Cell. Biol. 2005; 25:7092–7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Becerra S., Montes M., Hernández-Munain C., Sune C.. Prp40 pre-mRNA processing factor 40 homolog B (PRPF40B) associates with SF1 and U2AF65 and modulates alternative pre-mRNA splicing in vivo. RNA. 2015; 21:438–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morris D.P., Greenleaf A.L.. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2000; 275:39935–39943. [DOI] [PubMed] [Google Scholar]
- 82.Allen M., Friedler A., Schon O., Bycroft M.. The structure of an FF domain from human HYPA/FBP11. J. Mol. Biol. 2002; 323:411–416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.