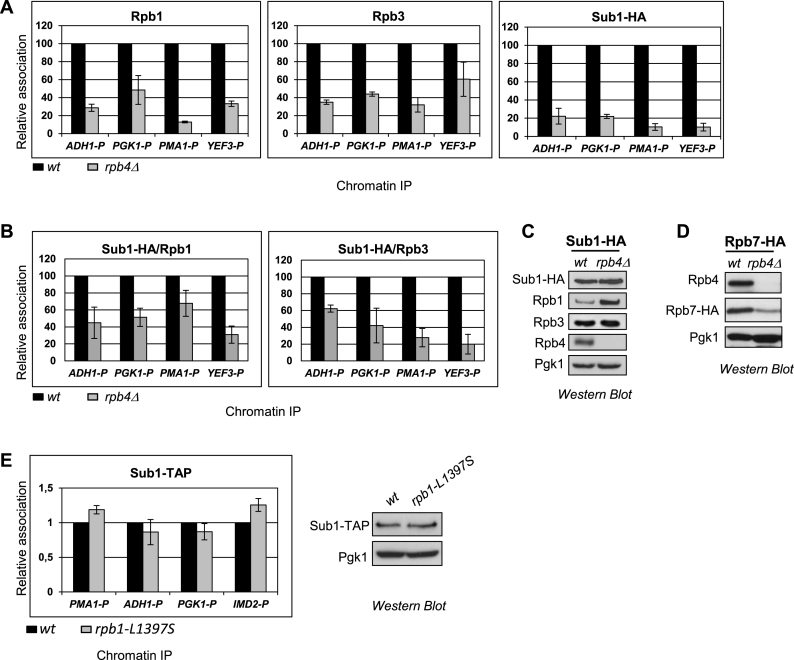
Figure 3.
A functional Rpb4/7 heterodimer is a requisite for Sub1 recruitment to gene promoters. (A) Chromatin immunoprecipitation (ChIP) analyses were performed using wt and rpb4Δ strains. Left panel: Rpb1 binding to the promoter (P) of four constitutively expressed genes, ADH1, PGK1, PMA1 and YEF3 was examined by qPCR. Results were quantified (see Materials and Methods), and relative Rpb1 binding in rpb4Δ cells is plotted relative to that in wt cells (set equal to 100). The data plotted correspond to mean values from at least three independent experiments, and the error bars represent standard deviations. Middle and Right panels show relative Rpb3 and Sub1–HA binding, respectively, plotted as for Rpb1. (B) Plots of Sub1–HA/Rpb1, Sub1–HA /Rpb3 ratios in wild-type (wt) and rpb4Δ using data from Figure 1A. (C) Whole cell extracts (WCE) were prepared from wild-type (wt) and rpb4Δ strains expressing Sub1–HA and analyzed by western blotting using the following antibodies: anti-HA, anti-Rpb1 (8W16G), anti-Rpb3, anti-Rpb4 (2Y14) and anti-Pgk1 as a control for total protein. (D) Whole cell extracts (WCE) were prepared from wild-type (wt) and rpb4Δ strains expressing Rpb7–HA and analyzed by Western blotting using anti-HA, anti-Rpb4 and anti-Pgk1. (E) Left panel: ChIP analysis in wt and rpb1-L1397S cells as in (A) to analyze Sub1–TAP occupancy at the promoter (P) of three constitutively expressed genes (PMA1, ADH1 and PGK1) and to the promoter of the IMD2 inducible gene, whose expression is regulated by Sub1 (38,82), and is upregulated in the rpb1-L1397S mutant (52). Right panel: WCE were prepared from wild-type (wt) and rpb1-L1397S strains expressing Sub1–TAP and analyzed by Western blotting using anti-TAP and anti-Pgk1 as loading control.