Figure 1.
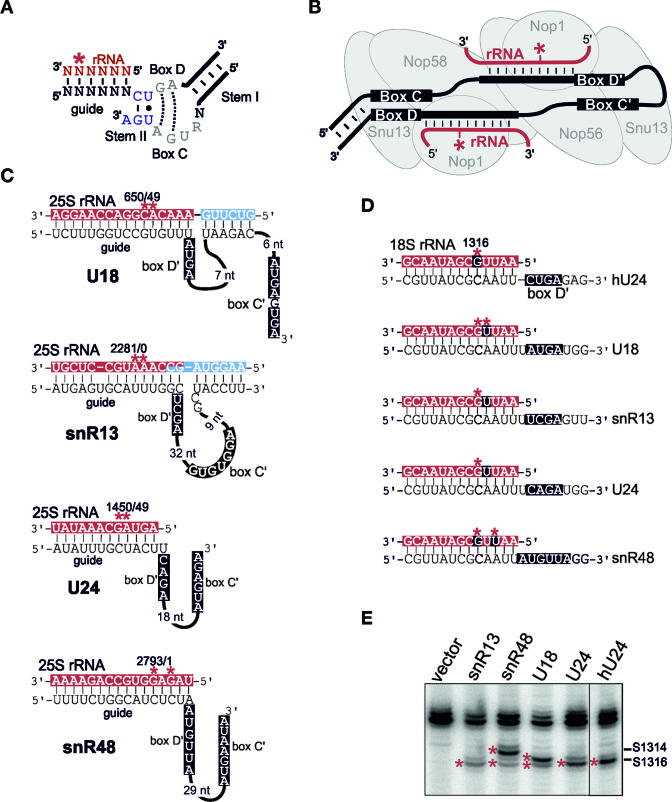
The 2΄-O-methylation activity of snR13, snR48, U18 and U24 C΄/D΄ motifs. (A) Schematic representation of the box C/D motif with the adjacent guide sequence base-paired to the rRNA target (shown in red with the target nucleotide indicated by an asterisk). The sequence of the C and D boxes are shown. The positions of stem I and II in the motif are indicated. Note that the C/D motif is rotated 180° relative to its orientation in (B) so that it is in the same orientation as the C΄/D΄ motif. (B) A schematic model of the box C/D snoRNP complex. The snoRNA is shown in black, with the C, D, C΄ and D΄ boxes indicated. The rRNA is shown in red, with the base-pairing interactions indicated. The asterisk represents the site modified. The box C/D snoRNP proteins are represented as grey filled ellipses. (C) rRNA (upper) and snoRNA (lower) sequences, with both conventional guide-rRNA interactions (rRNA shaded red) and novel extra (or accessory) base-pairing (rRNA shaded blue) interactions, for the S. cerevisiae box C/D snoRNAs that direct multi-site modification. Where sequences are shaded both red and blue, this indicates an overlap between the conventional and extra-base-pairing. The C΄ and D΄ sequences are shown in white with a black background. The guide sequences in the snoRNA are indicated. Note for snR48 there is no obvious D΄ box and the whole conserved area is marked (see later). The modified nucleotides are indicated by an asterisk together with the rRNA nucleotide number. (D) The guide sequence and D΄ motif of the artificial snoRNA constructs used to test the function of the various C΄/D΄ motifs is shown (the full sequence used is shown in Supplementary Figure S1B and C). The D΄ boxes are presented in white with a black background. The target 18S sequence (white text on red background) is shown with the expected (black background) and actual (asterisk) methylation sites indicated. (E) Constructs expressing artificial snoRNAs containing the C΄/D΄ motifs from U18, U24, snR13, snR48 and human (h)U24 (as indicated above each lane) were transformed into yeast cells. RNA was extracted from the cells and analysed by primer extension to detect rRNA methylation. The position of the stop corresponding to methylation of the target nucleotides, S1314 and S1316 in the 18S rRNA, are indicated on the left. Bands corresponding to 2΄-O-methylation are indicated by an asterisk. Northern blotting was used to control for snoRNA expression (Supplementary Figure S2A).