Figure 3.
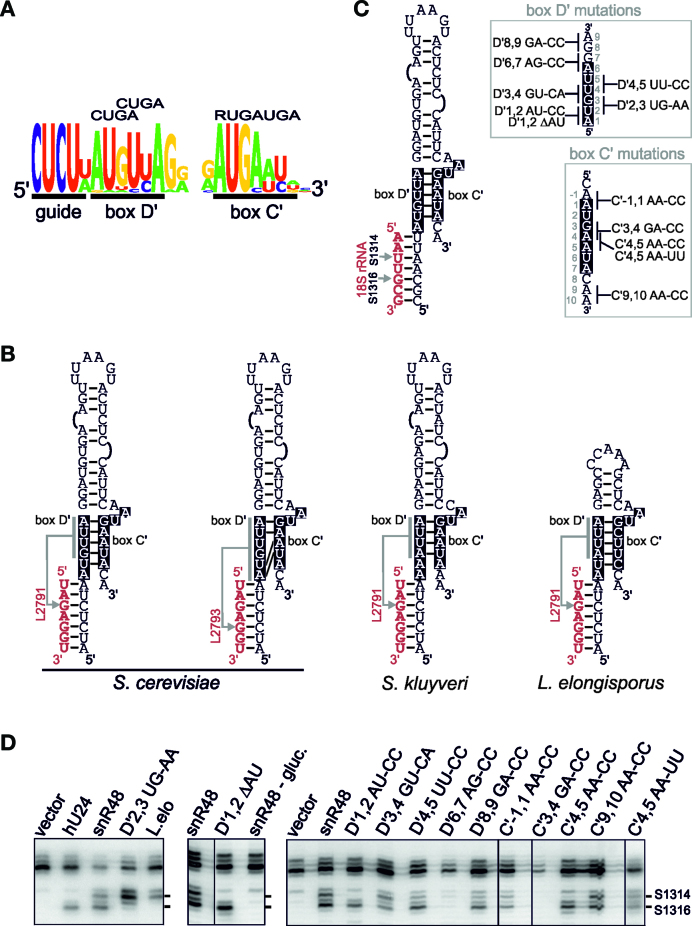
The snR48 C'D' motif is unique and directs 2΄-O-methylation at multiple sites in the target region. (A) A WebLogo representation of the conservation of the guide, D΄ box and C΄ box sequences of the snR48 snoRNA derived from the alignment of all the available yeast snR48 sequences (24). The consensus sequence for the C΄ and D΄ boxes is shown above the WebLogo image. Note, two potential alternative positions for the D΄ box are shown. The diagram was prepared using the WebLogo software (29). (B) Secondary structure of the snR48 box C΄/D΄ motif and guide sequence base-paired to the 25S rRNA from S. cerevisiae, S. kluyveri and L. elongisporus. Note two alternative structures are shown for the S. cerevisiae snR48. The C΄ and D΄ boxes are shown in white on a black background. Arrows indicate the site to be modified. (C) Secondary structure of the snR48 box C΄/D΄ motif, in the context of the artificial snoRNA targeting sites 1314 and 1316 in the 18S rRNA. The C΄ and D΄ boxes are shown in white on a black background. Arrows indicate the site to be modified. The mutations to the C΄ and D΄ boxes are shown to the right. (D) Constructs expressing artificial snoRNAs containing the wild-type and mutant snR48 C΄/D΄ motifs (as indicated above each lane) were transformed into yeast cells. RNA was extracted from the cells and analysed by primer extension to detect rRNA methylation. The position of the two target nucleotides (S1314 and S1316) is indicated on the right. L.elo is the artificial snoRNA with the C΄/D΄ motif from L. elongisporus snR48. snR48 glucose: yeast containing the plasmid encoding the artificial snoRNA with the snR48 C΄/D΄ motif, grown on glucose containing media. The levels of the various snoRNAs were determined by Northern blotting (Supplementary Figure S2C).