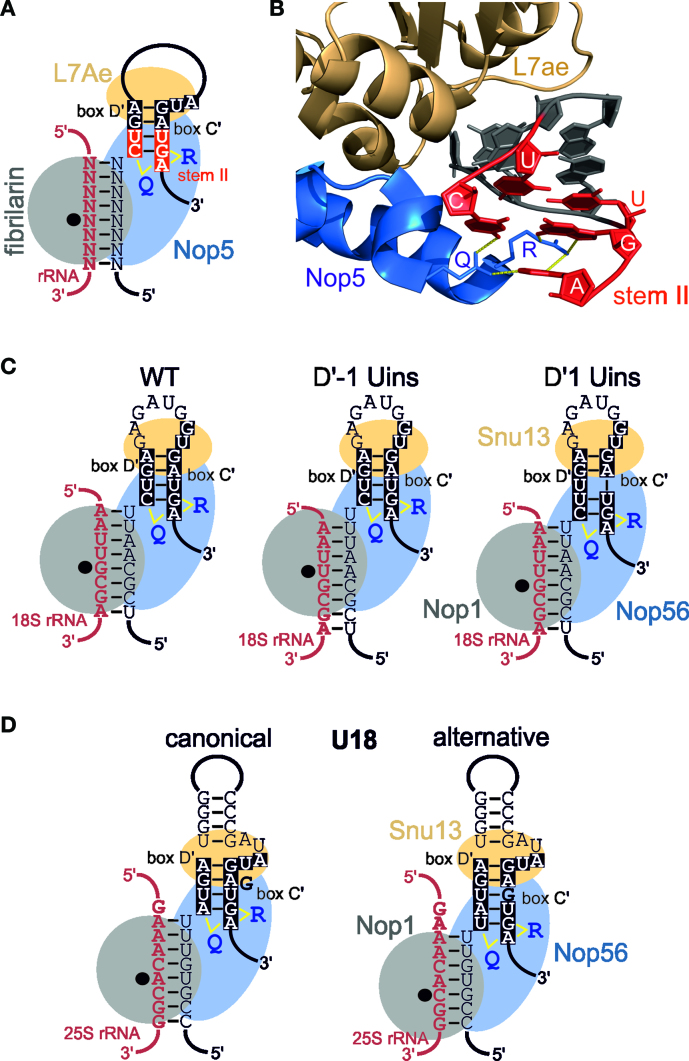
Figure 6.
Interaction of Nop56 with stem II of the C΄/D΄ motif dictates the site of 2΄-O-methylation. (A) Schematic model of the Archaeal C΄/D΄ motif and guide region base-paired to a target rRNA (shown in red). The box C/D snoRNP proteins are represented as grey filled ellipses. The black dot indicates the active site of fibrillarin (Nop1 in yeast) and is positioned over the nucleotide to be modified. The Q and R in Nop5 represent amino acids Q296 and R339 that make contact with stem II of both the C/D and C΄/D΄ motif motifs. Stem II is the C΄/D΄ motif is shown in white on a red background. The upper part of the C΄/D΄ motif, which is bound by L7Ae, is shown in white with a black background. (B) Recognition of stem II of the box C/D motif by the Nop domain of Archaeal Nop5 (22). Cartoon views of the relevant regions of Nop5 and L7Ae are shown. Amino acid side chains are only shown for Q296 and R339. The snoRNA is shown in cartoon form and only the C΄/D΄ motif is shown for clarity. Stem II is shown in red and the upper part, bound by L7Ae, is shown in dark grey. The identity of the nucleotides in stem II is indicated. Hydrogen bonds between Q296 and R339 I Nop5 and stem II of the C΄/D΄ motif are shown in yellow. (C) Organisation of the snoRNP proteins on the artificial snoRNA containing the hU24 C΄/D΄ motif and the D΄ box insertions presented in Figure 4. The schematic organisation is the same as in (A) except the whole C΄/D΄ motif is shown in white with a black background. Note the slight, one nucleotide shift, in the position of Nop56, and he corresponding change in Nop1 position, in the complex based on the recognition of stem II in the C΄/D΄ motif. (D) Organisation of the snoRNP proteins on the U18 snoRNA based on the canonical and alternative secondary structures presented in Figure 2B. The schematic organisation is the same as in (C).