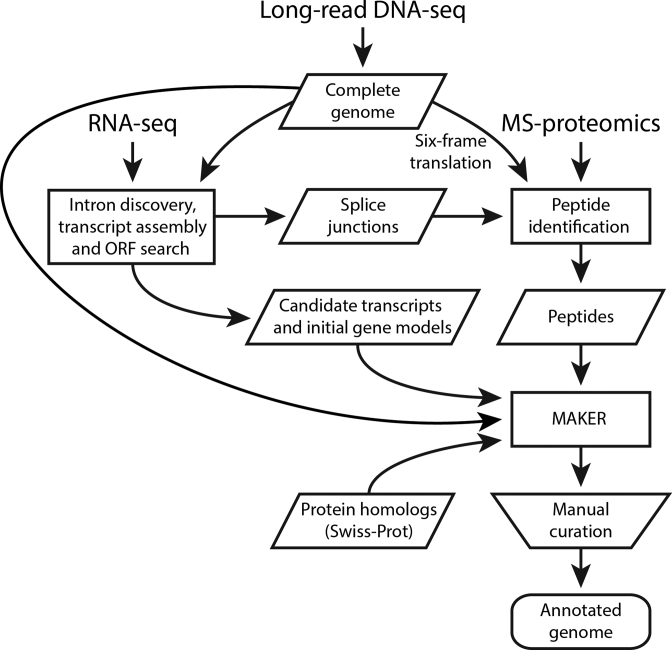
Figure 1.
Integrative genome annotation workflow. Data from four different sources (long-read DNA sequencing, RNA-seq, MS-based proteomics and Swiss-Prot reviewed proteins) were integrated using an evidence-based genome annotation framework (MAKER). Transcripts were assembled from RNA-seq reads using Trinity and PASA was used to identify likely protein-coding regions to provide gene models for initial gene predictions. Three ab initio gene predictors (GeneMark-ES, Augustus and SNAP) were included in MAKER. Augustus and SNAP were iteratively trained based on MAKER-generated gene models (see Materials and Methods and Supplementary Table S2). The computationally inferred gene structures were manually curated. Shapes are used according to workflow figure standards (rectangles show processes, data are in parallelograms, the trapezoid indicates a manual step and the rounded rectangle represents output).