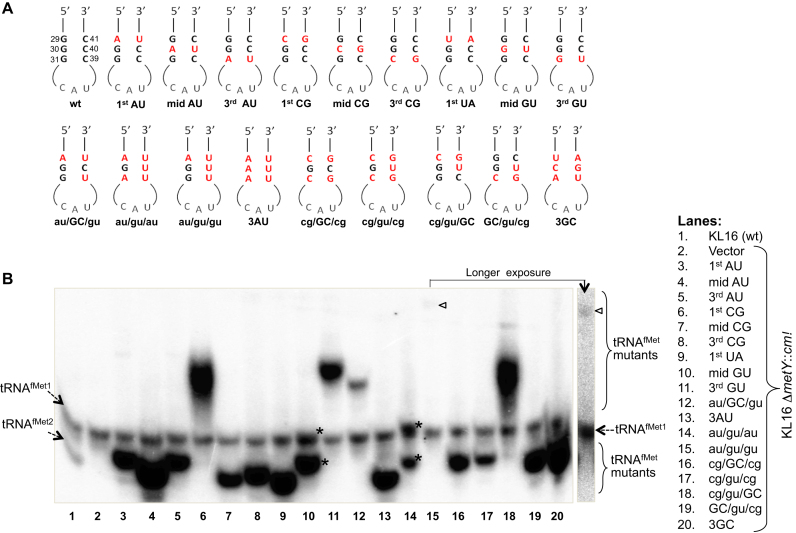
Figure 1.
Generation and analysis of 3GC mutants of tRNAfMet. (A) Sequences of the anticodon stem loops of the mutants used in this study. (B) Expression analysis of the mutant tRNAfMet. Approximately 10 μg of total tRNA preparations from E. coli KL16, or KL16ΔmetY::cm (retains tRNAfMet1, as indicated) harboring plasmids containing the mutant metY genes were separated on native polyacrylamide gel electrophoresis (PAGE), transferred onto nytran membrane and hybridized with D-loop ini tRNA probe (complementary to 2–27 nt position) common to all tRNAfMet. tRNAMet (detected by met elongator D loop probe) serves as internal loading control. The two conformer of mid GU and au/gu/au tRNAs are marked by asterisks (*), and the band corresponding to au/gu/gu has been indicated by an arrowhead. Longer exposure of lane 15 has also been shown.