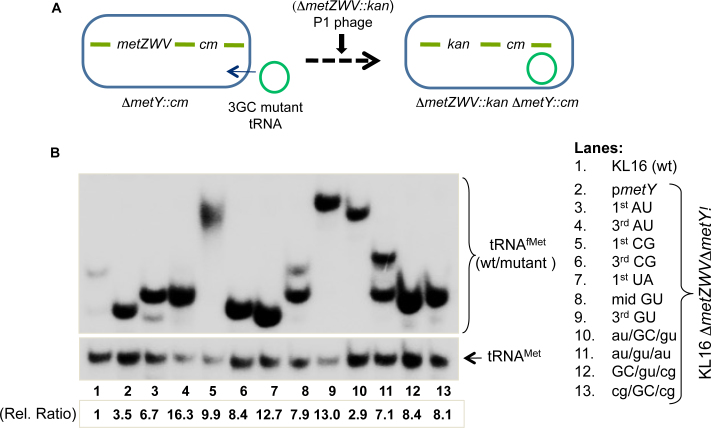
Figure 2.
Survival of E. coli on 3GC mutant tRNAfMet. (A) The ‘four-gene knockout’ strategy to check for the ability of mutant tRNAfMet to sustain E. coli. Among the two loci coding for tRNAfMet, the strain deleted for metY (ΔmetY::cm) was transformed with plasmid encoding mutant tRNAfMet; then the deletion of metZWV locus was attempted by P1 mediated transduction of ΔmetZWV::kan locus using Kan selection. (B) Northern blot analysis to confirm sustenance of the cell on the mutant tRNAfMet. Total tRNAs from cells sustained on wild-type or mutant metY genes were prepared and separated on native PAGE, transferred onto nytran membrane and probed with D-loop ini tRNA probe against tRNAfMet. For loading control, elongator tRNAMet was also probed with met elongator D loop probe, complementary to 2–27 nt position). Cellular abundances (relative ratios) of the initiator tRNAs in the strains were calculated relative to total tRNAfMet in wild-type strain (lane 1), after normalization with the corresponding tRNAMet.