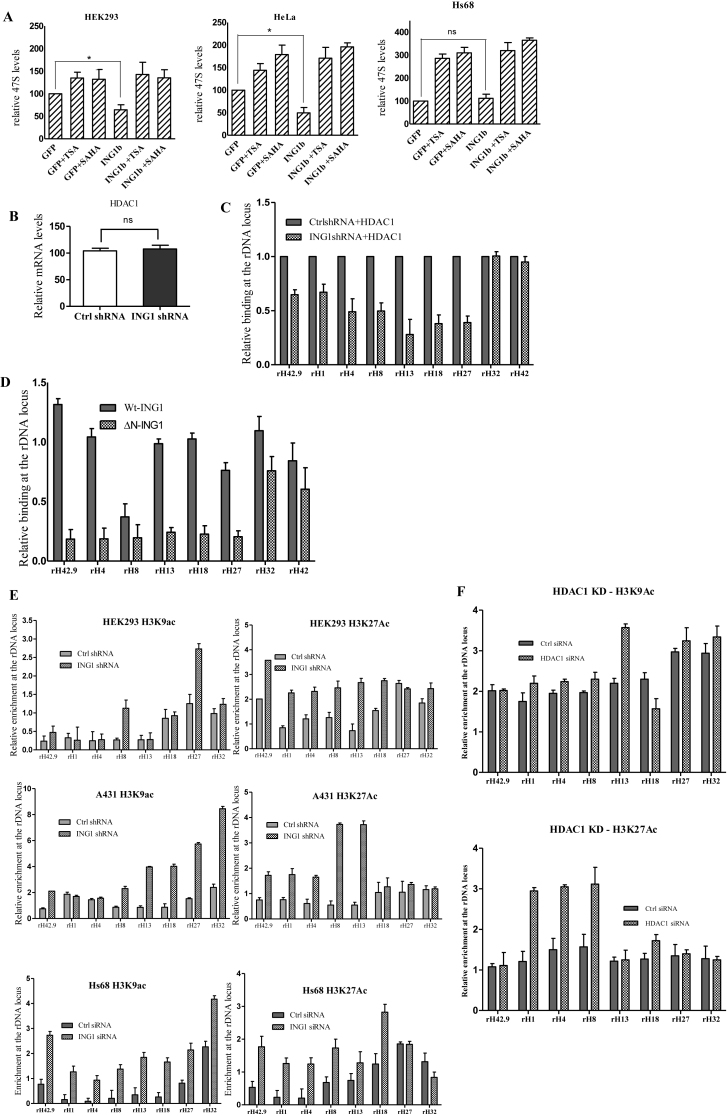
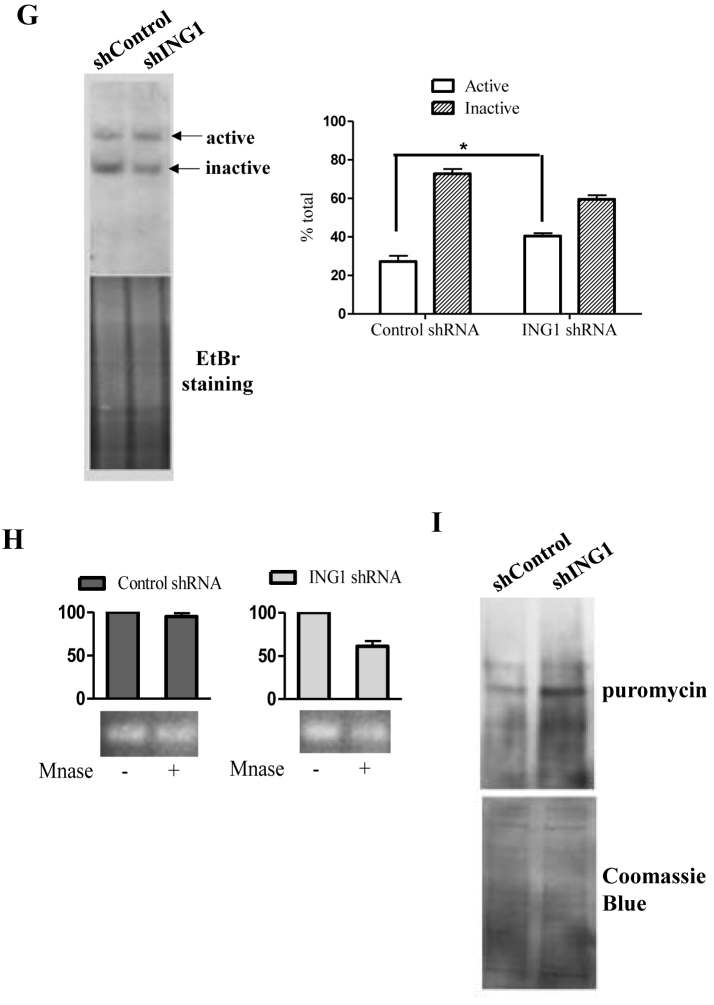
Figure 3.
ING1 affects the epigenetic state of rDNA. (A) HEK293 and HeLa cells transfected with control GFP or ING1 plasmid were treated with DMSO, TSA or SAHA for 24 h and checked for 47S rRNA levels by qRT-PCR (P < 0.05). Lower panel: Hs68 cells infected with Ad-GFP or Ad-ING1 for 24 h were treated with DMSO, SAHA or TSA. 47S rRNA levels were assayed using qRT-PCR (P < 0.05). (B) mRNA levels of HDAC1 in control and ING1 knockdown cells by qRT-PCR. (C) HEK293 cells treated with control or shING1 for 48 h were transfected with pcDNA-HDAC1-flag. Relative enrichment of HDAC1 was analyzed at the rDNA locus by ChIP (P < 0.05). (D) HEK293 cells transfected with pCI-Wt-ING1-flag or pCI-ING1ΔN-flag (lacks aa 1–125 constituting the SAID domain) were assayed for HDAC1 binding at rDNA by ChIP using HDAC1 antibody (P < 0.04). (E) HEK293 and A431 cells expressing shING1 and Hs68 cells transfected with control or siING1 were assayed by ChIP for relative enrichment of acetyl H3K9 and acetyl H3K27 at the rDNA locus. (F) Hs68 cells transfected with either control siRNA or HDAC1 siRNA were analysed for acetylation at H3K9 and K27 residues by ChIP assay (P < 0.05). (G) Nuclei of A431 cells stably expressing control or shING1 were crosslinked with psoralen and analyzed by biotin-labelled probe to the core rDNA promoter. The agarose gel stained with ethidium bromide serves as a loading control. The graph shows the means for quantification of active versus inactive rDNA repeats from three experiments (P < 0.05). (H) Nuclei from A431 control or shING1 cells were digested with MNase to estimate relative amounts of accessible rDNA repeats. Amplicons of the MNase digested samples were normalized with respect to undigested controls (P < 0.05). (I) A431 cells expressing control or shING1 were checked for puromycin incorporation as a measure of protein synthesis. The membrane stained with Coommassie Brilliant Blue serves as a loading control.