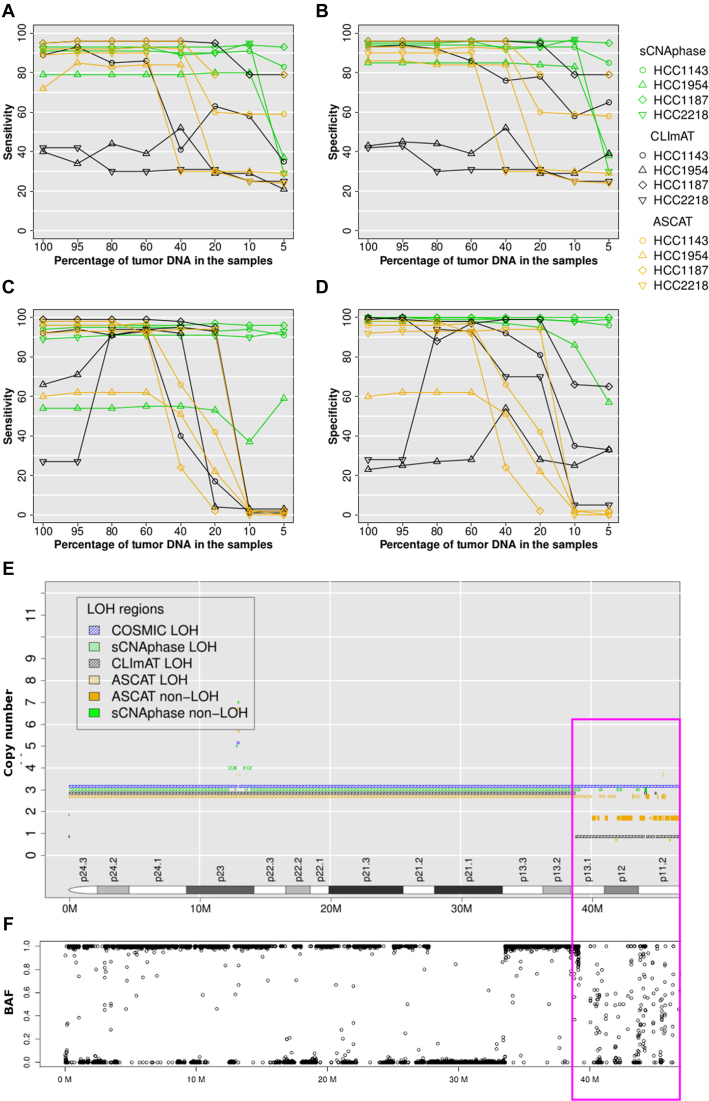
Figure 5.
sCNAphase recapitulates the COSMIC annotation of these cell-lines across a range of purities. (A and B) Consistency with COSMIC segmentation for copy number and (C and D) LOH at varied tumor purity. For tumor samples at different tumor purity, the base-pair sensitivity in (A and C) and base-pair specificity in (B and D), was calculated by overlapping sCNAphase, CLImAT and ASCAT segmentations estimated at a particular tumor purity with COSMIC segmentations based on 100% purity tumor samples. Each of the four cell-lines was represented a particular point shape as indicated in the legend. Results from sCNAphase, CLImAT and ASCAT are shown in green, black and orange, respectively. The copy number segmentations of chr9 p-arm HCC1187 from COSMIC, sCNAphase, CLImAT and ASCAT were shown in (E) in blue, green, black and orange hash lines, respectively. BAFs at this region in (F) support the loss of heterozygosity shown in COSMIC and CLImATsegmentation in p-arm except for the region highlighted in the pink box (39M-47M). For most of this 8M region, sCNAphase did not report copy number or LOH due to the highly variable BAFs that give merging errors (see Materials and Method).