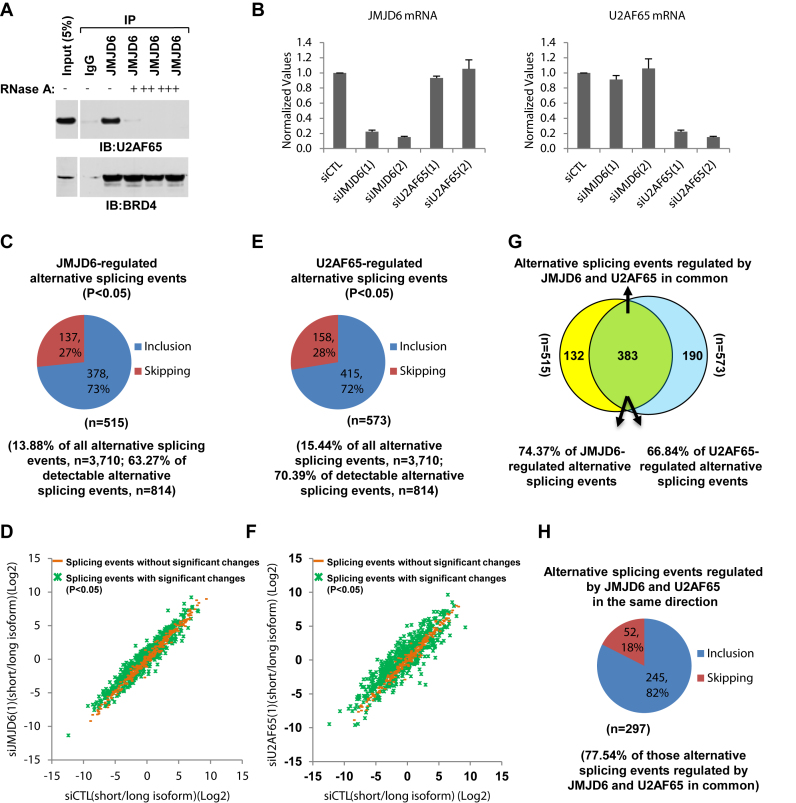
Figure 1.
JMJD6 and U2AF65 co-regulate alternative splicing. (A) JMJD6 interacted with U2AF65 in a RNA-dependent manner. Cell lysates from HEK293T cells were subjected to immunoprecipitation (IP) with control IgG or anti-JMJD6 antibody in the presence or absence of RNase A (+: 50 ng/μl; ++: 250 ng/μl; +++: 500 ng/μl) followed by immunoblotting (IB) with anti-U2AF65 (upper panel) or anti-BRD4 (bottom panel) antibodies. (B) HEK293T cells were transfected with control siRNA (siCTL) or two independent siRNAs specifically targeting JMJD6 (siJMJD6(1), siJMJD6(2)) or U2AF65 (siU2AF65(1), siU2AF65(2)) for 72 h followed by RNA extraction and RT-qPCR analysis using primers specifically targeting JMJD6 (left panel) or U2AF65 (right panel). Data shown was the relative fold change compared to control samples after normalization to actin. (C, E) Pie chart showing JMJD6- (C) or U2AF65- (E) regulated alternative splicing events, both exon inclusion and skipping, examined through RASL-Seq analysis using RNA samples described in (B) (P < 0.05). Data shown was from siJMJD6(1) (C) and siU2AF65(1) (E). (D, F) Scatter plot showing the isoform ratio (short versus long, log2) of all detectable alternative splicing events (read counts >5) in RASL-Seq when knocking down JMJD6 (D) or U2AF65 (F). (G) Venn diagram showing overlapping between JMJD6 and U2AF65-regulated alternative splicing events (P = 0.000785, hypergeometric test, expected background value: 52). (H) Pie chart showing alternative splicing events regulated by JMJD6 and U2AF65 in the same direction including exon inclusion and skipping (P < 0.05).