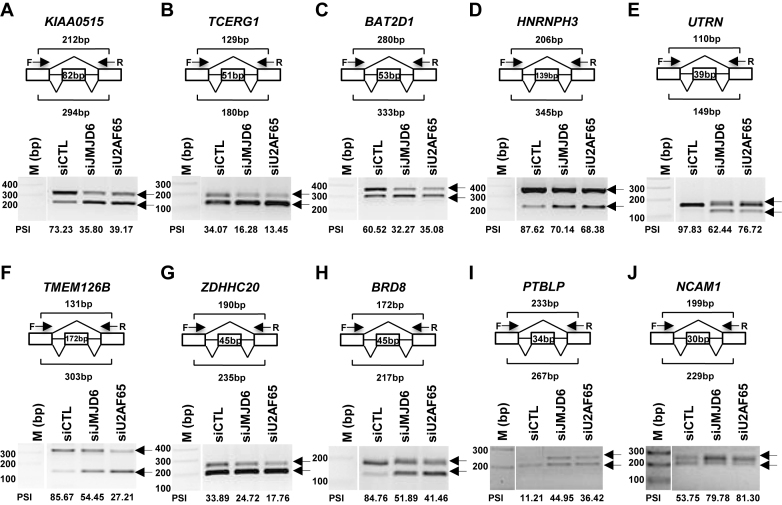
Figure 2.
Validation of alternative splicing events co-regulated by JMJD6 and U2AF65 through RT-PCR. (A–J) First-strand cDNA synthesis was done from RNA samples described in Figure 1B followed by standard PCR analysis (RT-PCR) using primer sets specifically targeting to the upstream and downstream exon relative to the cassette exon (alternatively spliced exon) to detect the expression of both short and long isoforms of representative genes as indicated. The length of the alternatively spliced exon as well as the expected length of the PCR products was shown as indicated. DNA marker (M) was included on the left. Percentage spliced in (PSI) was calculated as the ratio of the density of the long isoform versus that of the sum of the long and short isoforms. The position of the alternatively spliced exon in each gene was as follows: KIAA0515 (NM_013318, exon 30) (A); TCERG1 (BI091338, exon 3) (B); BAT2D1 (AV650960, exon 6) (C); HNRNPH3 (NM_012207, exon 3) (D); UTRN (NM_007124, exon 66) (E); TMEM126B (NM_018480, exon 2) (F); ZDHHC20 (uc001uod.1, exon 13) (G); BRD8 (NM_006696, exon 21) (H); PTBLP (NM_021190, exon 10) (I); NCAM1 (NM_181351, exon 9) (J). F: forward primer; R: reverse primer; bp: base pair.