Abstract
The genome of the cell is often exposed to DNA damaging agents and therefore requires an intricate well-regulated DNA damage response (DDR) to overcome its deleterious effects. The DDR needs proper regulation for its timely activation, repression, as well as appropriate choice of repair pathway. Studies in Saccharomyces cerevisiae have advanced our understanding of the DNA damage response, as well as the mechanisms the cell employs to maintain genome stability and how these mechanisms are regulated. Eukaryotic cells utilize post-translational modifications as a means for fine-tuning protein functions. Ubiquitylation and SUMOylation involve the attachment of small protein molecules onto proteins to modulate function or protein–protein interactions. SUMO in particular, was shown to act as a molecular glue when DNA damage occurs, facilitating the assembly of large protein complexes in repair foci. In other instances, SUMOylation alters a protein's biochemical activities, and interactions. SUMO-targeted ubiquitin ligases (STUbLs) are enzymes that target SUMOylated proteins for ubiquitylation and subsequent degradation, providing a function for the SUMO modification in the regulation and disassembly of repair complexes. Here, we discuss the major contributions of SUMO and STUbLs in the regulation of DNA damage repair pathways as well as in the maintenance of critical regions of the genome, namely rDNA regions, telomeres and the 2 μm circle in budding yeast.
INTRODUCTION
Protein functions within the cell need to be tightly controlled to ensure proper and timely function. This is achieved by numerous ways such as controlled expression, RNA and protein degradation, ligand binding as well as post-translational modifications. Covalent attachment of chemical groups to a protein provides a means for regulating protein functions as these attachments provide a quick reversible way of signaling and targeting that makes response to cellular changes rapid and dynamic. Phosphorylation, methylation and acetylation involve the attachment of small chemical groups on specific amino acid residues on proteins. This serves to either alter the protein surface property thus affect function, or act as a specific binding domain for other proteins thus modulate protein–protein interactions. Another post-translational modification is the addition of small proteins, ubiquitin and ubiquitin-like proteins such as small ubiquitin-like modifier (SUMO), through covalent attachment to the target protein, by the process of ubiquitylation and SUMOylation, respectively (1,2). Ubiquitin and SUMO are highly conserved within all eukaryotes and have been shown to play a critical role in most, if not all, cellular processes. Ubiquitylation and SUMOylation involve the addition of a single (mono) or a chain of molecules (poly) to the substrate protein. Poly-ubiquitylation of a protein has been most extensively studied as a means of signaling for proteasomal degradation by the 26S proteasomal system (3–5). However, it also plays an important role in signaling in various cellular pathways through altering protein–protein interactions (3). SUMO has also recently emerged as a critical factor in multiple biological processes such as DNA replication and transcription as well as the maintenance of genome integrity and the events following DNA damage (6). The ubiquitin and SUMO pathways share several similarities and common proteins, and are highly interconnected with SUMO-targeted ubiquitin ligases (STUbLs) providing the main link (7,8). These enzymes are recruited to SUMOylated proteins to subsequently ubiquitylate them leading in many cases to their proteasomal degradation.
One of the most important cellular processes, which in essence affects all other cellular processes, is protecting the genome. The cellular DNA is under continuous attack by DNA damaging agents, like ionizing radiation (IR), ultraviolet (UV) radiation, carcinogens, as well as endogenous stresses resulting from DNA replication errors and by-products of cellular metabolism such as reactive oxygen species. The capacity to deal with these DNA damaging agents is crucial for cell survival, and inefficiencies in DNA repair can lead to chromosomal aberrations and cancer in mammalian cells. When the cell encounters a DNA damaging agent, it activates an intricate response system, called the DNA damage response (DDR). This involves two interconnected pathways; one is to repair the DNA damage using several DNA repair pathways, and the other is to arrest the cell cycle in order to allow time for DNA repair and to prevent the propagation of damaged DNA into daughter cells (9,10). If the damage is too severe, cellular senescence or programmed cell death may occur. Both of these pathways are of critical importance in maintaining genome integrity and mutations in the proteins involved are frequent causes of carcinogenesis. Most of the basic mechanisms and factors involved in the DDR are well understood; however, what remains a mystery is how these pathways are regulated and the crosstalk that exists between them. One of these regulatory mechanisms is post-translational modifications on proteins involved in DDR, namely ubiquitylation and SUMOylation. Hence, ubiquitin, SUMO and the enzymes involved in their conjugation and processing are now regarded as critical players in maintaining genome stability. Studies in yeast Saccharomyces cerevisiae have played a key role in understanding the intricate process of DNA damage repair and the maintenance of genome stability. This review focuses on the role that SUMO and STUbLs play in the DDR in budding yeast and the maintenance of critical DNA elements, in particular rDNA, telomeres and the episomic 2 μm circle.
THE SUMOYLATION PROCESS AND COMPONENTS INVOLVED
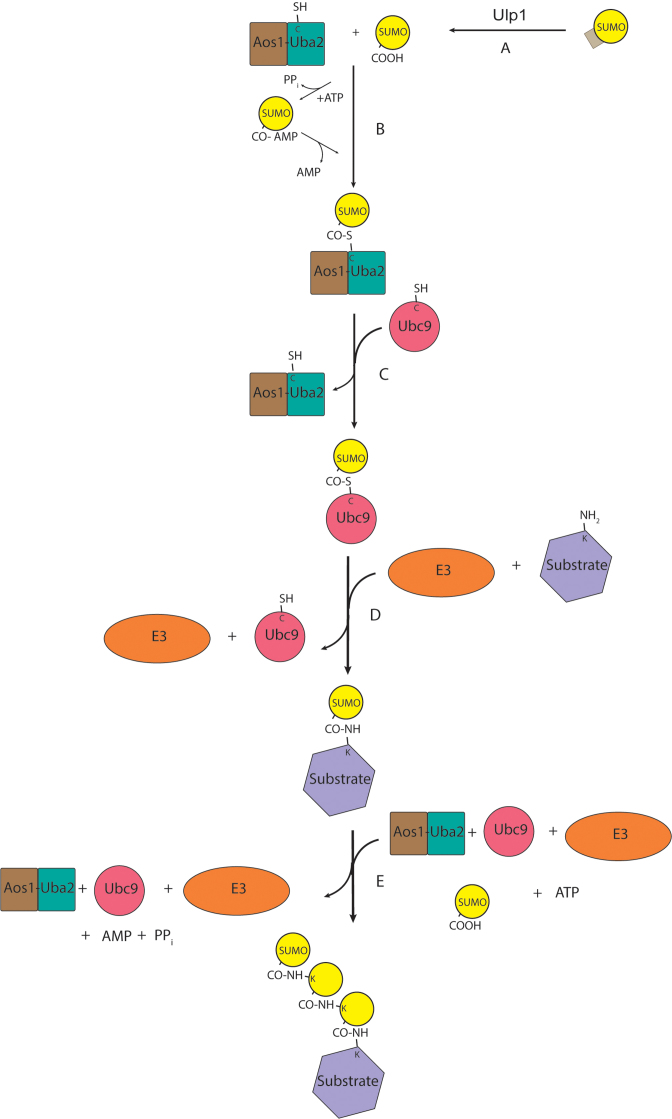
Although ubiquitin and SUMO only share 20% sequence identity, the conjugation of SUMO (Smt3 in S. cerevisiae) to proteins shows high resemblance to the ubiquitylation process [(8,11,12) and Figure 1]. The SUMOylation process also includes the action of E1-E2-E3 cascade of enzymes, and is conjugated to a large number of substrates. It involves an initial step of processing of Smt3 to expose a di-glycine residue at the C-terminus, followed by activation by the E1 activating enzyme complex, Aos1-Uba2 (13). Next is the conjugation to the E2 conjugating enzyme, Ubc9 and finally ligation to a substrate protein by a few E3 ligases including Siz1, Siz2, Mms21 (also called Nse2, a part of Smc5/6 complex) and Cst9 (meiosis specific E3) (14,15). The final ligation step of SUMOylation involves the formation of an isopeptide bond between the C-terminal glycine of Smt3 and an internal lysine in the protein. Unlike in the ubiquitin pathway, where E3 ligases act as the substrate receptors and recruit substrates for conjugation by E2s, the SUMO E2 enzyme Ubc9 can bind to substrates and catalyze the conjugation of SUMO to them directly without the need for SUMO E3s. However, SUMO E3 ligases confer higher selectivity to the process. Typically, SUMOylated sites are lysines within a consensus motif ΨKXE, where Ψ represents a large hydrophobic amino acid and X represents any amino acid (6). Other SUMOylated lysine sites, however, have also been reported (16,17). Similar to ubiquitin, SUMO can also be attached as a single moiety (mono-SUMOylation), as several moieties at multiple sites (multi-SUMOylation), or as a chain (poly-SUMOylation) (18). Poly-SUMO chains are attached through one of the three lysines in the N-terminus of SUMO (K11, K15 and K19 in Smt3). While SUMO is essential for yeast viability, poly-SUMOylation is not (19). The SUMO signal is removed by SUMO proteases, Ulp1 and Ulp2. Ulp1 is the major de-SUMOylating enzyme and is localized to the nuclear pores (20–22). Ulp1 is also responsible for the maturation of Smt3 to become a substrate for conjugation (23), and this accounts for the inviability of ulp1 null mutants (24). A mutation in the ULP1 gene results in an allele named nib1, which shows nibbled colony appearance and growth defects as a result of the hyperamplification of the 2 μm circle (25,26). Ulp2, on the other hand, is present throughout the nucleus and is specifically involved in de-SUMOylating poly-SUMOylated substrates (19,27,28). ulp2 null mutants are viable but show several growth defects (28). The conjugated SUMO moieties are recognized by two types of motifs; SUMO interacting motif (SIM) and Zn finger (ZZ) motif (29,30). The presence of tandem SIMs in a protein allows it to specifically bind poly-SUMOylated proteins (31).
Figure 1.
The SUMOylation process. It involves four steps; (A) Processing of small ubiquitin-like modifier (SUMO) by Ulp1 SUMO protease to expose a C-terminal di-glycine residue. (B) Activation; Aos1-Uba2 E1 activating enzyme uses the energy of ATP to form a SUMO-adenylyl intermediate followed by the conjugation of SUMO to a cysteine in E1 and the release of AMP. (C) Conjugation; Ubc9 E2 SUMO conjugating enzyme catalyze the transfer of SUMO from E1 to the active site cysteine of E2. (D) Ligation; Ubc9 or SUMO E3 ligases catalyzes the transfer of SUMO to the substrate through an isopeptide bond between a substrate lysine and the C-terminal glycine of SUMO. (E) Multiple rounds; the poly-SUMOylation process involves multiple rounds of SUMOylation onto one of the lysines on SUMO itself.
Unlike ubiquitylation, SUMOylation of target proteins does not serve as a signal for degradation. In fact, it has been shown to be involved in signaling in a large number of cellular processes, like nuclear transport, gene transcription, and DNA repair (16,17,29,32–35). Large scale SUMOylation of DNA repair proteins of all repair pathways occurs upon DNA damage (36), in a manner analogous to but independent of the phosphorylation network by checkpoint kinases in the DDR. This is known as DNA damage-induced SUMOylation (17,29,32–37). The SUMOylation process is very intriguing in that it includes a very small number of conjugating enzymes, and that the modified substrates represent a small fraction of the total substrates, yet the signal is transduced effectively. This was best explained when the protein group modification nature of the SUMO-conjugating system in response to a highly specific trigger was understood (37). The SUMOylation reaction does not target a specific substrate, but a group of proteins resulting in an additive or redundant effect (37). Interestingly, the SUMO and ubiquitin signals have been shown to occur on the same lysine residues leading to different outcomes, indicating possible competition between both pathways. The crosstalk between these pathways was further demonstrated by the identification of STUbLs, which are E3 ubiquitin ligases having SIMs and thus target SUMOylated proteins for ubiquitylation (7,38). STUbLs also have the ability to conjugate ubiquitin at the growing end of a SUMO chain, forming SUMO-ubiquitin hybrid chains, which may function to terminate the growing SUMO chain or to target the protein for proteasomal degradation as seen with Slx5-Slx8 STUbLs (7,18). The Slx5-Slx8 STUbL complex will be discussed in more detail in the section on the role of STUbLs in DSB repair. Proteasomal degradation by the 26S ubiquitin proteasome is highly conserved between species and is found in all eukaryotes. The 26S proteasome complex is responsible for degrading ubiquitin marked proteins, typically those that are poly-ubiquitylated using K48-linkage (3–5). The AAA ATPase Cdc48 also participates in the ubiquitin pathway as a chaperone protein to disassemble protein complexes and present the ubiquitylated proteins to the proteasome (39,40). This segregase activity requires Cdc48 to interact with cofactors, such as Ufd1, Npl4 and Doa1, to recognize the ubiquitylated substrates, and sometimes involves SUMO modification as well.
DNA DAMAGE REPAIR
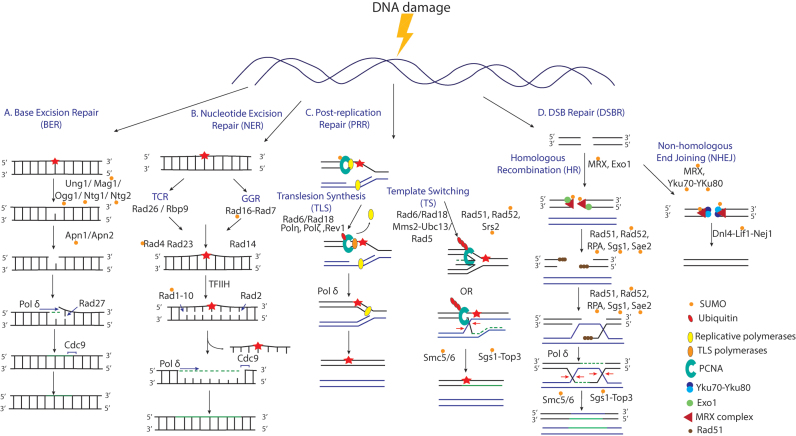
As discussed earlier, the genome of the cell is under continuous attack from DNA damaging agents that warrants for a complex DDR. Proteins involved include DNA repair factors such as nucleases, helicases, scaffold proteins and signaling factors. Interactions between repair factors need to be switched on and off in a rapid, reversible and dynamic manner in response to DNA damage. The conjugation of ubiquitin or SUMO moieties to repair proteins offers a dynamic way for their regulation. An overview of the DNA repair pathways is given in Figure 2, with the SUMOylated repair factors that have been analyzed in some detail marked. In the coming sections, we will provide a detailed account of the role of SUMO and STUbLs in the DDR as currently known. A summary of the effects of SUMOylation of some of the DNA repair proteins that have been analyzed in some detail is listed in Table 1.
Figure 2.
SUMO involvement in DNA repair pathways. (A) Base excision repair (BER); several DNA N-glycosylases were shown to be SUMOylated upon DNA damage, such as Ogg1, Ntg1, Ntg2 and Mag1, as well as the AP endonuclease, Apn1. (B) Nucleotide Excision Repair (NER); several NER proteins were found to be SUMOylated upon UV irradiation, such as Rad16, Rad7 and Rad4, as well as NER proteins that are also involved in other repair pathways, such as Rad1, Rad10, Rpb4 and Rad3. (C) Post-replication repair (PRR), in case a lesion is encountered by the DNA replication machinery, it can be bypassed by one of two sub-pathways, translesion synthesis (TLS) and template switching (TS). SUMO-PCNA acts as a prerequisite for template switching. Proteins involved in the resolution of sister chromatid junctions (SCJs), an intermediate in, T.S., were also shown to be SUMOylated, such as Smc5 and Sgs1. (D) DSB repair (DSB repair), proteins of the homologous recombination (HR) sub-pathway have been shown to be SUMOylated upon damage, such as RPA subunits, Rfa1 and Rfa2, MRX complex, Rad52, Rad59, Srs2, Sae2. This is in addition to non-homologous end joining (NHEJ) proteins, Yku70-Yku80 and Lif1. Table 1 lists the effects of the SUMOylation of the repair factors that were analyzed in some detail.
Table 1. The major SUMOylated proteins involved in DNA repair*.
| Substrate | Position | E3 | Function |
|---|---|---|---|
| Ntg1 | K364 | Nuclear localization of Ntg1 upon oxidative damage (42). | |
| Rad4 | Siz2 | Possibly promotes Rad4 functions as it accumulates in absence of downstream NER proteins (46). | |
| Rad1 | K32 | Siz1, Siz2 | Decreases Rad1 binding, facilitating its dissociation from DNA post-cleavage (47). |
| PCNA | K164 | Siz1 (70) | Enhances interaction with Rad18 through SIM. Allows the switch to the ubiquitin modification and activation of damage avoidance pathways (78). |
| Allows interaction with Srs2, to inhibit the extension of the D-loop thus limiting crossovers (85), as well as in protecting against Rad59-dependent polyubiquitylation dependent, gene conversion (86). | |||
| Mediates interaction with Elg1 subunit of alternative clamp loader leading to its unloading from DNA after completion of replication (74,75). | |||
| K127 | Siz2 (71) | Deters PCNA interactions as this modification with PIP box containing proteins, such as Eco1 and Rfc1, as it occurs at the interdomain connecting loop (77). | |
| Smc5 | Mms21 | Promotes the function of Smc5/6 complex in resolving X-shaped structures from TS events (88). Inhibiting recombination and resolving X-shaped structures at rDNA regions (151, 152). | |
| Rad52 | K43, K44 and K253 (or K10, K11 and K220) (101) | Siz2 (101) | Promotes Rad52 stability (101). Decreases ssDNA and dsDNA binding affinity and strand annealing activity (102). Enhances interaction with Rad51 (103). Inhibits unproductive and toxic Rad51 filaments and surpasses the need for Srs2 (104), possibly through recruiting Cdc48-Ufd1 segregase which dislodges Rad51-Rad52 from DNA preventing unnecessary recombination (103). Relocalizes Rad52 out of nucleolus (152). Under non-damaging conditions, favors direct repeat intrachromosomal recombination (101), and SSA (102). Under MMS-induced damage SUMO-Rad52 is required for interchromosomal recombination (100). SUMO-Rad52 is an in vitro substrate for STUbL Slx5-Slx8 complex (105). |
| Srs2 | K1081, K1089 and K1142 | Siz1, Siz2 | Important for the interaction with Rad51 through Rad51 C-terminal SIMs. SUMO-Srs2 increases recombination at rDNA regions (34). |
| Sae2 | K97 | Siz1, Siz2 | Facilitates Sae2 processing of complex DSBs and increase its solubility (113). |
| Mre11-Rad50-Xrs2 (MRX) | Enhances its resection activity and directs repair to HR (113). Mre11 SUMOylation enhances SUMOylation of downstream HR proteins (36). | ||
| Yku70 | K588, K591, K592, K596 and K597 | Enhances DNA binding (114). Promotes anchoring of telomeres to nuclear envelope (114). | |
| Lif1 | K301 | Siz1, Siz2 | Decreases ssDNA binding and self-association. Inhibits NHEJ at persistent DSBs (115). |
| Top2 | Siz1, Siz2 | Localizes at the rDNA loci (150). | |
| Smc1, Smc3 (Cohesin) Smc2 (Condensin) | Mms21 | Binding to 5S rDNA region (150). | |
| Cdc13 | K909 | Siz1, Siz2 | Inhibits telomerase mediated lengthening and mediates interaction with Stn1 (165). |
| Rap1 | K240 and K246 | Decreases NHEJ inhibition activity of Rap1. Targets it for ubiquitin mediated degradation by STUbL Uls1 (173). | |
| Sgs1 | K621 | Siz1, Siz2 | Promotes alternative lengthening of telomeres in telomerase-deficient cells by promoting telomere-telomere recombination (174). |
| Flp1 | K375 | Siz1, Siz2 | Limits the Flp1-dependent DNA damage on the 2 μm plasmid and the HR-dependent repair that results in hyperamplification of the 2 μm plasmid and eventually clonal lethality. The SUMO modification possibly targets Flp1 for Slx5-Slx8- dependent ubiquitylation and proteasomal degradation (106, 179). |
| Rep1 | K305, K315 and K328 | Allows proper association with STB locus in the 2 μm plasmid (184), thus the efficient partitioning of the plasmid in daughter cells. | |
| Rep2 | Several sites which may include: K42, K44, K92, K124, K130, K134, K146, K148, K149, K177, K208, K226 or K227. | Allows proper association with STB locus in the 2μm plasmid (184), thus the efficient partitioning of the plasmid in daughter cells. | |
*The list is not comprehensive and only presents examples where individual modifications have been analyzed in some detail.
SUMO IN BASE EXCISION REPAIR (BER)
BER is the primary repair pathway for small, non-helix distorting base lesions. The damaged base is first excised from the DNA by DNA N-glycosylases creating an abasic or apurinic/apyrimidinic (AP) site. This AP site is recognized by AP endonucleases, which nick the DNA backbone 5΄ to the lesion creating a 3΄-substrate for DNA polymerase (Polδ or ε) to fill, followed by DNA ligase (Cdc9) to seal the nick (10). For an overview on BER, see Figure 2. Additional information on BER can be found in several reviews (10,41).
Many proteins of the BER pathway were found to be SUMOylated upon DNA damage, such as the N-glycosylases Ogg1, Ntg1, Ntg2 and Mag1, as well as the AP endonuclease, Apn1 (36,42). Ntg1 and Ntg2 have similar functions but show different cellular localization (43,44). Under normal growth conditions as well as oxidative stress, Ntg1 localizes to both the nucleus and the mitochondria, while Ntg2 shows exclusive nuclear localization (42–44). The SUMOylation of the nuclear fraction of Ntg1 on K364 was found to increase 5-fold upon both nuclear and mitochondrial oxidative stress (42). This SUMOylation was found to be important for the nuclear relocalization of Ntg1 upon oxidative DNA damage, and for full oxidative damage resistance (42). Whether relocalization of SUMO-Ntg1 is due to increased nuclear transport, increased nuclear retention or both, is still unclear. The function of the SUMO modification of the rest of the BER enzymes also needs to be studied in greater detail to fully understand the role of SUMO in BER. Nevertheless, SUMOylation of Ntg1 provides an example of how SUMOylation affects relocalization of a DNA repair protein, and is thus crucial for conferring cellular survival following oxidative stress.
SUMO IN NUCLEOTIDE EXCISION REPAIR (NER)
NER is the primary repair pathway responsible for repair of bulky DNA lesions that cause distortion of the DNA helix such as cyclobutane pyrimidine dimers resulting from UV damage. These lesions are recognized by Rad4-Rad23 and Rad14 repair factors, followed by unwinding of DNA by TFIIH and Rad3 helicases, together with Mms19 and RPA. Next is the excision of an oligonucleotide fragment of about 24–27 nucleotides around the DNA lesion by the action of endonucleases, Rad2 and Rad1-Rad10 (10). The lesion recognition step divides NER into two pathways: transcription coupled repair (TCR) and global genome repair (GGR). TCR pathway repairs lesions encountered by RNA Polymerase II (RNA Pol II) causing it to stall, and thus repairs lesions on the template strand. TCR requires the additional function of either Rad26 or the Rpb9 subunit of RNA Pol II for damage detection. GGR can repair damage on template and non-template strands, and involves the Rad16-Rad7 complex. For an overview on NER, see Figure 2. Additional information on NER can be found in several reviews (10,45).
Enzymes of the SUMO pathway have been implicated in NER as seen with the sensitivity of Δsiz1Δsiz2 double mutants to UV damage (46). Genetic data suggest that Siz1 and Siz2 act in both the Rad16-dependent GGR and the Rpb9 sub-pathway of TCR, with a minimum role in the Rad26-branch of TCR (46). Several NER proteins were found to be SUMOylated upon UV irradiation, such as Rad16, Rad7 and Rad4, as well as NER proteins that are also involved in other repair pathways, such as Rad1, Rad10, Rpb4 and Rad3 (16,36). SUMOylation of Rad16 was found not to affect NER (46,47). Rad4 was shown to be SUMOylated by Siz2 and accumulates in the SUMOylated form when any of the downstream NER proteins, such as Rad33, Rad1 and Rad14, are absent (46). This suggests a role for SUMOylation in the function of Rad4 that remains to be identified. Rad1, a subunit of the NEF1 complex that includes Rad10 and Rad14, is also SUMOylated upon UV irradiation (47). Rad1 is SUMOylated on K32 by both Siz1 and Siz2 in a manner dependent on the upstream processing of the lesion by the NER machinery and the loading of Rad1 on the damaged DNA (47). Slx4 scaffold protein has been shown to be important for SUMOylation of Rad1 (47), in addition to its role in activating Rad1 nucleolytic activity after its recruitment to 3΄-flaps (48). SUMOylation of Rad1, however, did not affect its nucleolytic activity or interactions with other repair proteins (47). Nonetheless, cells expressing non-SUMOylatable Rad1-K32R showed sensitivity to high doses of camptothecin and UV irradiation, but not to a single cut induced by HO endonuclease (47). This, together with the finding that SUMO-Rad1 showed less DNA binding affinity (49), suggests that SUMOylation of Rad1 facilitates its dissociation from DNA post-cleavage, allowing Rad1 to handle the large amount of lesions that occur at high doses of camptothecin and UV (47). It would be interesting to understand how SUMOylation affects the DNA binding affinity of Rad1 and whether it is perhaps by inducing a conformational change in its DNA binding domain.
Complex DNA lesions such as DNA–protein crosslinks (DPCs) and protein–protein adducts can be produced enzymatically as an intermediate step in some DNA processes such as topoisomerase–DNA intermediate complexes, and non-enzymatically by some kinds of DNA damaging agents such as formaldehyde (50). Small DPCs can be resolved and repaired by NER, e.g. camptothecin-stalled Top1 DNA cleavage complexes that are resolved by tyrosyl–DNA phosphodiesterase I (Tdp1) (51). In the absence of Tdp1 or in the case of larger DPCs, the SUMO system is involved. The dual acting SUMO-ligase–protease, Wss1, has been recently implicated in the resolution of DPCs. Wss1 was originally identified as a metalloprotease from the family of minigluzicins (52). Its protease activity, however, was found to be latent and only activated upon DNA damage, and upon the binding and oligomerization of Wss1 on ssDNA lesions (53). Under normal conditions and when first recruited to DNA damage, Wss1 catalyzes the formation of poly-SUMO chains and is thus considered a SUMO-ligase (53). Polymeric SUMO possibly leads to further recruitment of Wss1 at damaged sites, its oligomerization and the activation of its proteolytic activities (53). At sites of damage, Wss1 was found to form a ternary complex with Cdc48/Doa1 and acts to proteolytically cleave and disassemble proteins from the damaged sites, to target them to the vacuole for processing (50,53). This process is thought to reduce the size of the DNA–protein adduct, so that they can be processed by the NER machinery. Whether this process involves any ubiquitin modification which Cdc48 normally targets, or whether Cdc48 cofactors Ufd1/Npl4 are involved in the subsequent steps are still unclear. However, this provides an additional involvement for SUMO in DNA repair, through the processing of complex DNA structures and targeting them for vacuolar autophagy.
SUMO IN POST-REPLICATION REPAIR (PRR)
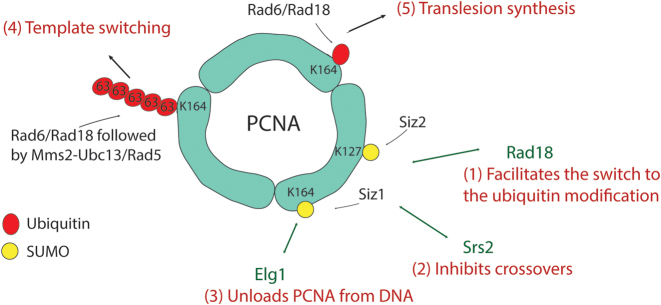
PRR is the pathway that allows the bypass of a DNA lesion when encountered by the replication machinery. It involves two sub-pathways: the error-prone translesion synthesis (TLS) and the error-free template switching (TS). PRR primarily involves proteins of the Rad6 epistasis group, nearly half of which are ubiquitin conjugating enzymes (E2s) and ubiquitin ligases (E3s) (54,55). The PRR pathway is mostly regulated by ubiquitin and SUMO modifications on the sliding clamp of the replicative machinery; proliferating cell nuclear antigen (PCNA) or Pol30. PCNA not only interacts with DNA polymerase as part of the replisome, but also interacts with proteins involved in the downstream processing of the newly synthesized DNA, such as nucleosome assembly and sister chromatid cohesion, as well as with DNA repair proteins at stalled replication forks. These interactions are mediated through direct interaction with hydrophobic regions in the PCNA through PIP/PIM (PCNA-interacting protein/motif) box or after PCNA modification with ubiquitin and SUMO. PCNA has 18 lysines that can be modified, however, the most frequently ubiquitylated and SUMOylated site is K164 [(56) and Figure 3]. For an overview on PRR, see Figure 2. Additional information on PRR can be found in several reviews (57–59).
Figure 3.
SUMO as a regulator of PCNA ubiquitylation and repair pathway choice. (1) SUMOylation on K164 by Siz1 or K127 by Siz2 mediates interaction with Rad18 through Rad18 SIM, thus switching to the ubiquitin modification upon DNA damage. (2) SUMOylation of PCNA also serves to recruit Srs2, leading to inhibition of Polδ/η limiting D-loop extension and crossovers. (3) SUMOylation of PCNA also facilitates its interaction with Elg1 alternative clamp loader, to unload PCNA from DNA upon completion of DNA synthesis. (4) K63 linked poly-ubiquitylation on K164 is mediated by Mms2-Ubc13/Rad5 and requires the prior mono-ubiquitylation by Rad6/Rad18. It results in template switching through either fork regression or SCJs. (5) Ubiquitylation of K164 of PCNA is mediated by Rad6/Rad18 and leads to translesion synthesis by recruiting TLS polymerases to the damage site.
Stalled replication forks accumulate RPA-bound ssDNA which recruits Rad18 (E3 ubiquitin ligase), and together with Rad6 (E2 ubiquitin conjugating enzyme), they mono-ubiquitylate PCNA on K164 activating the TLS sub-pathway. This signals the recruitment of TLS polymerases such as Rev1, Polη and Polζ, which can incorporate nucleotides correctly or incorrectly opposite the damaged one (60–63). Further ubiquitylation of PCNA by Ubc13-Mms2/Rad5 (E2/E3) enzymes (17,60) activates the error-free sub-pathway, T.S., which involves the use of the newly replicated sister chromatid as a template to accurately copy past the lesion. TS is proposed to occur through two mechanisms, either fork reversal creating a chicken foot like structure (64,65), or through recombinational invasion of the sister chromatid involving proteins of the homologous recombination (HR) pathway (66,67). The accumulation of sister chromatid junctions (SCJs) in cells impaired for their resolution, supports a recombinational mode of TS (17,33). HR also operates as a salvage pathway in PRR (67,68). Although, this pathway was termed post-replication repair, the belief was that it happens coupled to the replication fork. Several recent studies, however, have shown that PRR mostly occurs after bulk replication has occurred, in late S/G2 phase (69). Both modes of PRR are considered DNA damage tolerance or damage avoidance pathways, which allows the replication machinery to bypass the damage and the lesion to be repaired at a later time (67). The choice and regulation of the pathways involved are currently being extensively studied and have been shown to be at least partly regulated by SUMOylation of PCNA. Figure 3 shows the modifications of PCNA and their effects on the choice of PRR sub-pathway.
SUMOylation of PCNA occurs constitutively in the S-phase typically at K164 by Siz1 (70), and less efficiently at K127 by Siz2 (71). Despite possible competition between the ubiquitin and SUMO modifications on K164, it has been shown that SUMOylation of PCNA does not affect ubiquitylation levels (72), supposedly because both modifications can occur on different subunits of the same PCNA homotrimer (17,73). SUMOylation of PCNA particularly on K164 occurs when it is loaded onto the DNA (71), and enhances its interaction with the alternative clamp loader Elg1, facilitating its unloading from DNA after the completion of DNA synthesis and ligation (74,75). Consistent with this, SUMOylation of PCNA was increased in cdc9-1 mutants that harbor un-ligated Okazaki fragments, likely reflecting the retention of PCNA on the DNA (76). Elg1 dependent unloading of SUMO-PCNA may also serve to remove the SUMO-PCNA signal when recombination is required (74). PCNA SUMOylation on K127, on the other hand, deters PCNA interactions with PIP/PIM box containing proteins, like Eco1 and Rfc1, as this modification occurs at the interdomain-connecting loop of PCNA (77).
SUMOylation of PCNA has been shown to play a role in the maintenance of genome stability through at least two mechanisms (Figure 3). First, SUMO-PCNA directly facilitates its interaction with Rad18-SIM, which shows particular preference for SUMOylated PCNA (78) and thus promotes the switch to the damage-induced ubiquitin modification (33,79). The existence of SIM on the Rad18 ubiquitin ligase enhancing its substrate targeting depicts Rad18 as a STUbL (80). Second, SUMOylated PCNA allows its interaction with the SIM of the anti-recombinase helicase Srs2, which is known for its activity in dislodging unproductive Rad51 nucleofilaments keeping HR in check and in promoting poly-ubiquitin dependent PRR, both of which rely on its helicase activity (81–83). Despite the presumption that SUMO-PCNA recruits Srs2 to primarily dislodge Rad51 nucleofilaments at stalled forks, this Srs2 activity was shown not to depend on its interaction with SUMO-PCNA, as it was maintained in a Srs2 mutant that is unable to interact with SUMO-PCNA (84). In fact, Srs2 when complexed with SUMO-PCNA decreases the Srs2 that is available for dismantling Rad51 filaments (85), as well as precludes the SUMO modification of Srs2 itself (49), which is important for interacting with Rad51 (34). The genomic instability and increase in gene conversion rates observed when SUMO-PCNA Srs2 interaction is inhibited may actually occur due to the loss of another function they play, which is the blocking of Polδ/η mediated extension of the recombination intermediates generated during the repair of stalled forks (85). Crossovers are associated with the formation of long D-loops and therefore limiting D-loop extension restricts gene conversion events at stalled forks. A recent study, in fact, showed that SUMO-PCNA protects against Rad59-dependent gene conversion events that are mediated by Mms2-Ubc13/Rad5-dependent polyubiquitylation of PCNA, thus directly implicating SUMO-PCNA as a regulator of the downstream template switching events (86). Two Srs2 activities may contribute to the control of gene conversion, the helicase activity that promotes branch migration and synthesis dependent strand annealing (SDSA), and the inhibition of DNA repair synthesis when complexed with SUMO-PCNA. The inhibition of DNA repair synthesis resulting from the interaction between Srs2 and SUMO-PCNA underlies the sensitivity of Δrad18 mutants to, U.V., as the alternative pathways required for survival in these cells are inhibited. This also explains the partial rescue of Δrad18 mutants when Srs2 SUMO-PCNA interaction is inhibited (49,85). Altogether, SUMOylation of PCNA allows the switch to the ubiquitin modification upon DNA damage to commit the repair to the damage avoidance pathways, as well as regulate the poly-ubiquitylation mediated template switching events of PRR through limiting D-loop extension and cross overs.
The accumulation of SCJs in SUMO deficient mutants shows the importance of SUMO in resolving them. Mms21 SUMO ligase, which is part of the Smc5/6 complex, has been shown to be important in resolving Rad18-dependent SCJs, as well as HR-mediated SCJs that may operate as a salvage pathway in PRR. Smc5/6 is one of the structural maintenance of chromosomes (SMC) complexes that include cohesin (Smc1/3) and condensin (Smc2/4), that are responsible for sister chromatid cohesion and chromosome condensation, respectively (87). The Smc5/6 complex has been shown to play a role in resolving DNA-mediated linkages and has been proposed to be named ‘resolvin’ (87). Δsgs1, Δtop3 and mms21-11 single mutants accumulate Rad18-dependent X-shaped structures, indicating the importance of the Smc5/6 complex in the resolution of recombination intermediates (33). These results suggest that in a strain having functional SUMOylation activity, Rad18-PRR is the predominant error-free damage avoidance pathway, and that Mms21-dependent SUMOylation contributes to the resolution of SCJs formed during template switching. Among the Mms21 SUMOylation substrates that may contribute to SCJ accumulation are Smc5 and Sgs1 (33,88,89). The Smc5/6 complex is also responsible for SUMOylating components of the replisome, such as Mcm2 and the Pol2 subunit of Polε (90), and this SUMOylation is important for replication fork progression in the presence of DNA-damaging agents.
SUMO IN DOUBLE STRAND BREAK (DSB) REPAIR
DSBs, in which both strands of the DNA are broken, can be repaired by two main pathways. The first is simple re-ligation of the broken ends through non-homologous end joining (NHEJ), which occurs throughout the cell cycle. NHEJ can be error-prone, as loss of nucleotides around the break can lead to mutagenesis. Lesion recognition during NHEJ depends on the MRX (Mre11-Rad50-Xrs2) complex and the Yku70-Yku80 heterodimer. The Yku70-Yku80 complex binds the broken ends and mediates their ligation by the DNA Ligase IV complex (Dnl4-Lif1-Nej1) (91). The second pathway to repair DSBs is HR, which involves the use of the sister chromatid as a template to copy past the break. HR can only occur after DNA is replicated during S phase, after the sister chromatid becomes available. HR involves the concerted action of various repair factors. The MRX complex first binds to the broken ends, and together with Sae2 remove aberrant DNA end structures, and resect 50–100 nucleotides at the 5΄-end forming a 3΄-overhang (92). Further resection is achieved by the action of two nucleases, the Exo1 exonuclease and the Dna2 5΄-flap endonuclease, which partners with the Sgs1-Top3-Rmi3 helicase-topoisomerase complex. This creates long 3΄-ssDNA tails on which the ssDNA binding protein RPA binds. RPA is then displaced by Rad51, forming a nucleoprotein filament that together with Rad52, invade the sister chromatid forming a D-loop in search for homologous regions. Once found, Rad51 dissociates from the synaptic region and DNA polymerases Polδ or Polε extend at the 3΄-end of the invading strand (93). The second end of the DSB can be captured to form double Holliday junctions, where their resolution could result in crossovers or non-crossovers. Helicases such as Sgs1, Mph1 and Srs2 promote strand displacement through a sub-pathway called SDSA, which is preferred during mitosis and results in non-crossovers. If the second end of the DSB is lost, break-induced replication (BIR) can occur, where the D-loop turns into a replication fork copying the entire chromosome arm, as seen in alternative lengthening of critically short telomeres in telomerase-deficient cells (93). DSBs between repeats can be repaired by single strand annealing (SSA), where DNA end resection reveals homologous regions on the same strand. These homologous regions anneal to each other through the action of Rad52 and Rad59, resulting in the loss of the middle region. SSA and some types of BIR are Rad51-independent and Rad59-dependent (93). For an overview on DSB repair, see Figure 2. Additional information on DSB repair can be found in several reviews (10,93–95).
As discussed earlier, large-scale SUMOylation of DNA repair proteins occurs in response to DNA damage as part of the DDR (36,37). This is evident following DSBs, where the proteins involved in DSB processing and subsequent repair are highly SUMOylated in response to damaging agents, and SUMO-deficient strains exhibit high sensitivity to DSB inducing agents (36,37). The SUMOylation wave involves proteins of both NHEJ and HR pathways of DSB repair and is catalyzed by Siz2 in response to ssDNA exposure by Exo1 nuclease and Sgs1 helicase at DSBs (37). The interactions necessary to concentrate Siz2 at DSBs are not fully understood. The SAP (SAF-A/B, Acinus and PIAS) domain of Siz2 is composed of a core (cSAP) and an α-helical (eSAP) region, and is essential for protein and DNA binding (96,97). The cSAP domain of Siz2 was initially identified as the one responsible for general DNA binding, and the C-terminal SIM domain responsible for the interaction with SUMOylated Mre11, which is mediated by Ubc9 at MMS-induced DSBs, both allowing the recruitment of Siz2 to DSB sites (37). The dependence of the SUMOylation of many HR proteins on Mre11 was shown by the pronounced decrease in their SUMOylation in Δmre11 mutants (37). Recently, however, it was shown that the eSAP domain is responsible for Siz2 interaction with RPA, recruiting Siz2 to DSBs, similar to how the inducer of the checkpoint phosphorylation wave Mec1 kinase is recruited to DSBs (98,99). The SUMOylation wave has been shown to enhance interactions between the repair proteins through the multiple SIMs that they harbor, to presumably facilitate the assembly of repair foci (37). Almost all HR proteins are SUMOylated in response to DSB damage, including the RPA subunits Rfa1 and Rfa2, subunits of the MRX complex, Rad50 and Xrs2, Rad52, Rad59, Srs2 and Sae2 (36,37). The known effects of the modification of these proteins are listed in Table 1. The best studied examples are Rad52 and Srs2, and will be discussed with more detail below.
The MMS-induced SUMOylation of Rad52 was found to primarily occur when cells are entering the S-phase, but not when blocked in the G1 or G2 phases (100). The SUMOylation of Rad52 depends on the MRX complex (37,101), and is stimulated by Rad52 binding to ssDNA, whether or not RPA-bound (102). SUMO-Rad52 shows less ssDNA and dsDNA binding affinity and less strand annealing activity than unmodified Rad52 (102). SUMOylation of Rad52 results in both pro- and anti-recombinational effects. The pro-recombinational effects of SUMO-Rad52 are shown by the reduction of interchromosomal recombination to approximately half in cells expressing non-SUMOylatable Rad52 (Rad52-3KR) upon MMS-induced DNA damage, but not upon a single HO-induced cut (100). While it remains plausible that the tested mutant has some impaired activity that is required upon damage, the pro-recombinational effects can be attributed to the possibility that Rad52 SUMOylation facilitates its interaction with Rad51 C-terminal SIM (103) in order to facilitate the loading of Rad51 onto ssDNA. Under non-damaging conditions, SUMO-Rad52 seems to affect repair choice, favoring direct repeat intrachromosomal recombination (101) and SSA (102). However, since the SUMO signal is primarily induced upon DNA damage, it is possible that the observed effects do not reflect an actual preference, but rather an assay-biased effect. On the other hand, SUMO-Rad52 helps prevent superfluous recombination possibly through recruitment of Cdc48-Ufd1 segregase, which harbors SIMs, to remove improperly loaded Rad51-Rad52 from DNA (103, 104). SUMO-Rad52 was proven to be an in vitro substrate for the STUbL complex Slx5-Slx8, which polyubiquitylates SUMOylated substrates and targets them for degradation (105). This has also been demonstrated in vivo through the accumulation of flourescent Rad52 foci at the nucleolus (106), as well as increased ChIP enrichment of Rad52 at CAG-130 repeats (107), in Δslx5 and Δslx8 mutants (Figure 4B). Additionally, rad52-3KR, Δslx5 and Δslx8 single mutants exhibit expansions and contractions at CAG repeats (107) and nucleolar regions (106), typical of hyper-recombination at these regions in the absence of the SUMO regulatory signal (107). At the cellular level, however, neither Δslx5 nor Δslx8 mutants displayed slower degradation or accumulation of SUMO-Rad52 (106). In fact, less SUMOylated Rad52 and several other HR proteins were observed in Δslx8 and Δslx5 mutants upon MMS-induced DNA damage (106). The contradictory results on the accumulation of Rad52 in Δslx5 and Δslx8 mutants may be explained by the defined localization of the SUMO signal that may not be reflected on the cellular level of the protein. All in all, SUMOylation of Rad52 appears to occur after its binding to ssDNA, to facilitate its interaction with Rad51. SUMO-Rad52 decreases its DNA binding and recruits Cdc48 complex, both may allow the dissociation of Rad52-Rad51 complex as a means for recycling to handle the large amount of damage, or inhibit unneeded recombination particularly when cells are entering the S phase and the sister chromatid is not available. Under non-damaging conditions, particularly in the nucleolus and at CAG-repeats, SUMO-Rad52 primarily works to recruit Cdc48 segregase and to target Rad52 for Slx5-Slx8 mediated proteasomal degradation, thus inhibiting unnecessary recombination.
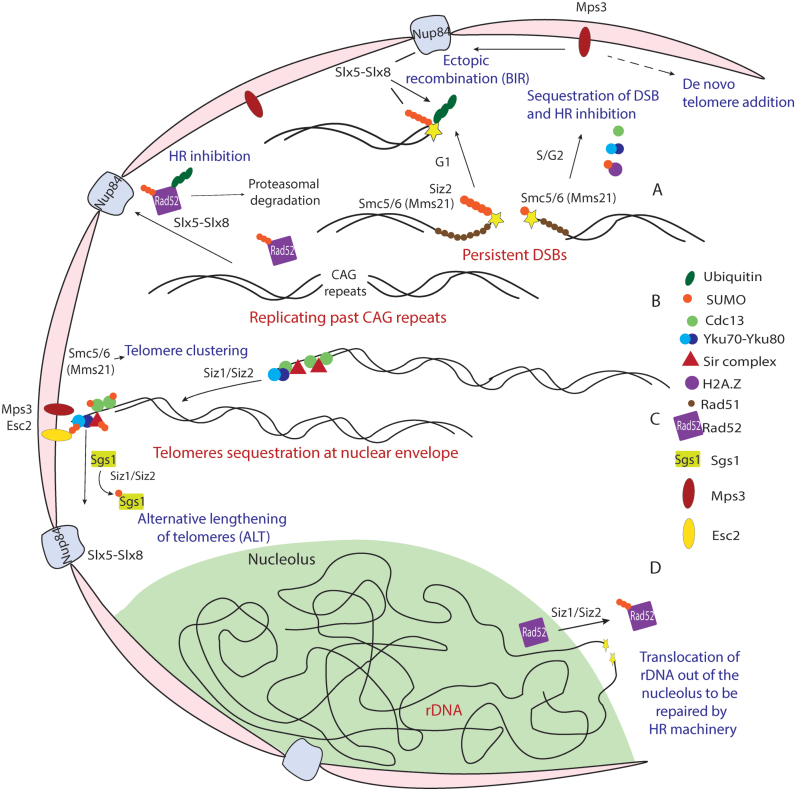
Figure 4.
Role of SUMO and STUbLs in nuclear sub-compartmentalization of DNA damage. (A) Persistent DSBs are translocated to the nuclear envelope in S/G2 phase, depending on mono-SUMOylation by Mms21 subunit of Smc5/6 complex, recruitment of SUMO-H2AZ, Rad51 and telomere proteins Cdc13 and Yku70-Yku80. This results in inhibition of recombination and possibly de novo telomere addition. Translocation to nuclear pores can occur in G1 phase depending on poly-SUMOylation requiring the sequential activity of Mms21 and Siz2. (B) Difficult to replicate repeat regions are translocated to the nuclear pores. The localization of Slx5-Slx8 at nuclear pores allows the ubiquitin dependent degradation of SUMOylated Rad52, thus serving to inhibit recombination at these stalled replication forks. (C) Telomere anchoring at the nuclear envelope inhibits recombination and requires the SUMOylation of the telomere associated proteins Cdc13, Yku70-Yku80 and the Sir complex. Smc5/6 complex is also required for telomere clustering. The SUMOylation of Sgs1 in telomerase-deficient cells translocates the telomeres to the nuclear pores, where alternative lengthening of telomeres by recombination can occur. (D) The highly repeat rich rDNA is located in the nucleolus sub-compartment. SUMOylation of Rad52 is required for its exclusion from the nucleolus, which is essential for inhibiting recombination at these regions. A DSB induced at rDNA requires its translocation outside of the nuclear sub-compartment to be repaired by the nuclear Rad52 pool.
Another interesting protein that gets SUMOylated is the Srs2 helicase. Srs2 has helicase and translocase activities and has been shown to possess both anti- and pro-recombinational roles in vivo. The multiple roles of Srs2 in DNA repair entail proper regulation of its function. Srs2 possesses a SIM and a PCNA interacting motif (PIM or PIP). The anti-recombinational role of Srs2 is mediated by both its SIM and PIM, as it is recruited to stalled replication forks by SUMO-PCNA to inhibit D-loop extension limiting crossovers. In addition, Srs2 dismantles Rad51-nucleofilaments through its translocase activity, inhibiting unwarranted HR (70,83,108,109). The pro-recombinational role of Srs2 depends on its helicase activity, where it promotes SDSA, which involves branch migration and non-crossover products (110). Srs2 also promotes Rad51-dependent and -independent recombination, particularly when phosphorylated by Cdk1 (Cdc28) (49,111,112). SUMOylation of Srs2 is mostly dependent on Ubc9 without the need for Siz1 or Siz2. The interaction between Srs2 and SUMO-charged Ubc9 is mediated by Srs2-SIM, thus SUMO-PCNA-Srs2 interaction inhibits SUMOylation of Srs2 by outcompeting Ubc9 (49). Cells expressing non-SUMOylatable Srs2 (srs2-3KR mutants) do not show altered intrachromosomal or interhomolog recombination rates but show increased recombination at rDNA regions (34). The SIM domain of Srs2 is important for mediating the pro-recombinational role of Srs2 and thus depends on the PCNA-unbound pool of Srs2 (49). The SUMOylation and SIM of Srs2 mediate its interaction with other HR proteins such as Rad51, Rad52, Mre11 and to a lesser extent Rad59 (34). Both the SIMs of Srs2 and Mre11 are important for their interaction, while in the case of the interaction with Rad52, only the SIM of Srs2 is essential (34). Rad51-Srs2 interaction depends on both SUMOylation and SIM of Srs2 as well as on the two SIMs at the C-terminus of Rad51 (34). All of these interactions are enhanced in srs2ΔPIM strains, indicating the competition between PCNA and HR proteins in binding Srs2 and thus regulating its recombinational role (34). In summary, PIM and SIM of Srs2 promote its anti-recombination activity by recruiting it to stalled replication forks by SUMO-PCNA. Whereas, in the absence of SUMO-PCNA interaction, SUMO and SIM of Srs2 mediate its interaction with HR proteins to promote recombination. These findings show how SUMOylation coordinates the pro- and anti-recombination activities of Srs2 and how it can modulate sometimes opposing functions of a protein.
Sarangi et al. recently studied the effect of SUMOylation of Sae2 nuclease and the MRX complex (113). They showed that SUMOylation of Sae2 requires both Siz1 and Siz2 and occurs on K97 (113). SUMOylation of Sae2 was found to facilitate the processing of complex DSBs such as hairpin-ends and those bound with Top1 following camptothecin treatment. SUMOylation of Sae2 was also found to increase its solubility, similar to its phosphorylation by Mec1 upon entering S phase in the presence of damage (113). SUMOylation of the MRX complex was also found to enhance the resection activities and direct the repair to HR (113). Collectively, the effects of SUMOylation of HR proteins indicate a role for SUMO in regulating HR repair upon DSBs.
NHEJ proteins, such as the Yku70, Yku80 and Lif1 (part of the DNA IV ligase complex), have also been shown to get SUMOylated upon DSB induced damage. Yku70 gets SUMOylated upon DNA damage induction through treatment with zeocin or other replication blocking agents, and requires prior binding to DNA and interaction with Yku80 (114). SUMOylation of Yku70 has been shown to stimulate NHEJ through enhancing its DNA binding (114) and to also affect its role in telomere maintenance, which will be discussed further in the coming sections. Lif1, another SUMOylated NHEJ protein, was shown to be SUMOylated at K301 at a basal level and induced upon DNA damage in a non-cell-cycle dependent manner (115). Unlike Rad52, binding of Lif1 to ssDNA inhibits its SUMOylation (115). Lif1 SUMOylation decreases its ssDNA binding activity and its self-association without affecting its interaction with Nej1, Xrs2 or Dnl4 (115). SUMOylation of Lif1 was shown to result in inhibition of NHEJ particularly at persistant DSBs (115).
SUMO-TARGETED UBIQUITIN LIGASES (STUBLS) IN DSB REPAIR AND PERINUCLEAR LOCALIZATION
The ubiquitin and SUMO pathways converge to regulate DSB repair as seen in the involvement of the STUbLs Slx5-Slx8 in recombinational repair. Slx5 and Slx8 were initially identified in screens for genes required for viability in sgs1 null mutants and displayed synthetic lethality (gene x), highlighting their role in recombinational repair (116). Slx5-Slx8 is a heterodimeric complex, which consists of the Slx8 RING finger E3 ligase that interacts with Ubc4 E2 enzyme and catalyzes the conjugation of ubiquitin to substrates. Slx5 harbors multiple SIMs, and was shown to be specifically targeted to poly-SUMOylated substrates to mediate their ubiquitylation and subsequent degradation (105,117,118). On account of this, a general hyper-poly-SUMOylation is observed in Δslx5 and Δslx8 mutants (18). The involvement of Slx5-Slx8 in DNA repair was further demonstrated by an increase in gross chromosomal rearrangements in Δslx5 and Δslx8 mutants (119). The involvement of the Slx5-Slx8 complex in the SUMO pathway was highlighted by the growth defects associated with hyperamplification of the 2 μm circle in Δslx5 and Δslx8 mutants that is typical of SUMO pathway mutants (117,120). These growth defects are dependent on proteins of the Rad51-independent recombinational repair (120). The role of SUMO in maintaining the 2 μm circle levels will be discussed later.
Slx5 and Slx8 have been shown to localize to nuclear pores (121), rDNA regions (106) and replication forks as indicated by the co-localization with PCNA (106). As previously discussed, Slx5-Slx8 seem to play an inhibitory role in recombination partly through SUMO-Rad52 degradation, particularly at nucleolar and repeat regions (106). An increase in mutation rates was observed in Δslx5 and Δslx8 mutants, as well as higher levels of Rad51-dependent and -independent recombination (106,119). This is in addition to increased Ddc2 and Rad52 foci and higher levels of Rad53 phosphorylation indicative of checkpoint activation in Δslx5 and Δslx8 mutants (106,119). Recently, the Slx5-Slx8 complex has been shown to play a role in repressing spontaneous and HU-induced Sgs1 foci, thus inhibiting unnecessary recombination (122). While the overall protein levels were shown to be unaffected (122), Slx5-Slx8 may function to target the neighboring pool of Sgs1 to proteasomal degradation, not affecting the overall levels. To answer this, the SUMOylation of Sgs1 in relation to spontaneous or HU-induced replication stalls in the absence of Slx5-Slx8, needs to be tested. Altogether, these findings suggest that the Slx5-Slx8 complex inhibits HR during replication and at repeat rich regions presumably through ubiquitylation and subsequent degradation of multiple SUMOylated factors that normally promote HR, thus keeping unnecessary HR in check.
A recently identified intriguing SUMO-related phenomenon is the relocalization of recalcitrant DSBs to the nuclear periphery (123,124), in a manner similar to telomere and rDNA nuclear membrane anchoring and the relocalization of actively transcribed genes to the nuclear pores [(125–128), Figure 4A]. Links between the SUMO pathway and the nuclear organization have long been suggested by the localization of the Ulp1 protease and the Slx5-Slx8 STUbL complex at the nuclear pores (126,129,130). Ulp1 localization at nuclear pores is mediated through interaction with the inner pore basket proteins Mlp1 and Mlp2, and is crucial for nuclear transport and genome stability (129). Slowly repaired DSBs are tethered to the inner nuclear envelope and this has been shown to inhibit ectopic recombination, thus preventing gross chromosomal rearrangements resulting from collapsed forks or unrepaired DSBs (123,124). This involves the recruitment of components of the telomerase machinery like Cdc13, Est1, Est2 and Yku70-Yku80 to DSBs to mediate the interaction with the Mps3 envelope protein (123,124). Recruitment to the nuclear periphery and interaction with the telomerase machinery can, but not necessarily will, result in de novo telomere addition during cell adaptation with unrepaired DSBs (131). The relocalization to the nuclear periphery has been suggested to involve the histone variant H2A.Z (Htz1), where it gets deposited around DSB sites early after DSB induction and subsequently SUMOylated (132). SUMO-H2A.Z, Rad51 and checkpoint activation were all shown to be important factors for the localization of DSBs to nuclear periphery [(132), Figure 4A]. However, since H2A.Z is important for Mps3 localization at the nuclear envelope (133), further studies are warranted to understand whether the defect in DSB relocation in Δhtz1 mutants is in fact due to impaired nuclear envelope proteins assembly. Other reports have shown association of persistant DSBs to nuclear pores in a manner dependent on Slx5-Slx8, the nuclear pore Nup84 complex and Mec1/Tel1 kinases (121). To reconcile the interaction of unrepaired DSBs with both the nuclear envelope and the nuclear pore, some suggested a sequential shuttling from the nuclear envelope to the nuclear pores (134), reminiscent of the alternative lengthening of telomeres that occurs in telomerase-deficient cells (in PML bodies in human cells), which involves proteins of the Rad51-independent recombination repair pathway [(107,135), Figure 4A].
Horigome et al. recently clarified that the choice of the association of persistent DSBs with either the nuclear pore or the nuclear envelope at least partly depends on the cell cycle stage and the extent of SUMOylation (136). They demonstrated that in S/G2 phase, mono-SUMOylation mediated by the Smc5/6-Mms21 complex results in association with the Mps3 nuclear envelope protein and inhibition of recombinational repair [(136), Figure 4A]. In the G1 phase, however, DSBs are directed to the nuclear pores through poly-SUMOylation mediated by the sequential activities of Mms21 and Siz2, leading to the recruitment of Slx5-Slx8 to the sites of damage to direct DSBs to nuclear pores [(136), Figure 4A]. The Slx5-Slx8 mediated relocation of DSBs to nuclear pores was shown to not entirely depend on the ubiquitin ligase activity of Slx8, but rather on the interaction of Slx5 with poly-SUMO chains, the Nse5 subunit of the Smc5/6 complex, and with the nuclear pore complex, Nup84 (136). At the pores, Slx5-Slx8 appears to ubiquitylate targets at the DSB site, mediating nuclear pore association that favors ectopic BIR and imprecise end joining (136). Intriguingly, the nucleoplasmic domain of Mps3 is required for the genome instability observed in Δslx5 strains (123), suggesting a loss of balance in the regulation of recombination between nuclear pores and envelope. In all cases, SUMO and its chain length play a major importance in the perinuclear localization of DSBs.
Collapsed replication forks at trinucleotide repeats, on the other hand, have been shown to interact only with nuclear pore proteins such as Nup84, but not with Mps3 [(107), Figure 4B]. At these difficult to replicate regions, Slx5-Slx8 association with Nup84 serves to target Rad52 for degradation, preventing contractions and expansions resulting from recombinational repair at these regions (107). Together, these results indicate a role for SUMOylation in DNA damage adaptation and repair pathway choice. It also indicates a role for Slx5-Slx8 at nuclear pores to inhibit de novo telomere addition, and depending on the type and region of damage, either mediate Rad51-independent recombination to enable cell adaptation or inhibit Rad52 recombinational events allowing the maintenance of genome stability (107,124).
Two putative STUbLs, Uls1 and Irc20, belonging to the Swi2/Snf2 family of ATPases, were also shown to be involved in DNA repair (137–139). Uls1 was originally identified to play a role in antagonizing silencing during mating type switching (140). Uls1 harbors a Snf2 helicase domain and an ubiquitin ligase RING finger domain that allows it to interact with the ubiquitin conjugating enzyme Ubc4 (118). Uls1 also possesses multiple SIMs similar to Slx5-Slx8 (118), suggesting a role in ubiquitylation of SUMOylated substrates; although this biochemical activity has not been demonstrated yet. It was initially assumed that Uls1 functions in a redundant pathway to Slx5-Slx8 (118). Recent physical and genetic evidence, however, are suggesting that Uls1 actually antagonizes Slx5 activity without affecting Slx5 and Slx8 protein levels or cellular localization (141). Uls1 has been shown to be important for S-phase progression in the presence of DNA damage, particularly in the Δrad52 mutant (139). Owing to its translocase activity, Uls1 has been shown to remove Rad51 nucleofilaments and inhibit unneeded recombination, particularly in strains lacking Rdh54 that predominantly removes Rad51 depositions on dsDNA (142). Genetic interactions with HR proteins suggest that Uls1 works upstream of Sgs1 possibly to facilitate Sgs1 activity (143), in a manner dependent on its helicase domain, and in a pathway independent on Mus81 and Yen1 nucleases as well as Srs2 and Mph1 helicases (139,143).
Irc20 was initially identified in a screen for gene deletions that caused increased recombination centers (Irc20), as shown by increased spontaneous fluorescent Rad52 foci, suggesting its role in regulating HR (137). Similar to Rad5 and Uls1, Irc20 has a Snf2 helicase domain in addition to a RING E3 domain (144,145) and was shown to play a role in transcriptional regulation dependent on both domains (146,147). Irc20 was shown to be an ubiquitin ligase in vitro (146), however, its in vivo substrates are still unknown. Irc20 also interacts with Cdc48 segregase and SUMO through at least two SIMs, in addition to being SUMOylated itself (146). Irc20 is also important for promoting SDSA, inhibiting crossovers and maintaining precise NHEJ, as seen by the defects in Δirc20 mutants (138). What the function of the helicase domain of Irc20 is, and which activity of Irc20 is responsible for the DNA repair defects observed in Δirc20 mutants, remain to be determined. Altogether, despite clear evidence that Slx5-Slx8, Uls1 and Irc20 STUbLs are involved in DSB repair and replication stress response, their molecular targets are still not identified.
SUMO AND STUBLS IN THE MAINTENANCE OF CRITICAL DNA REGIONS
Certain DNA sequences in the genome such as the repeat-rich rDNA regions in the nucleolus or telomeres require proper maintenance. Replication past rDNA regions is particularly challenging as repetitive sequences commonly form secondary structures that stall the replication fork and could lead to fork collapse (107). Repair of damage at these regions requires special care to avoid expansions and contractions of the rDNA repeats, which commonly occurs during recombinational repair between repeats. Telomeres, on the other hand, require protection from being recognized as DSBs by the repair machinery, which could result in telomere fusions (148). SUMOylation and STUbLs have been shown to play an important role in maintaining the integrity of these special DNA structures.
rDNA regions
In S. cerevisiae, rDNA is composed of 100–200 tandem repeats encoding the 35S and 5S ribosomal RNA. The importance of SUMOylation in rDNA maintenance is highlighted by the accumulation of fluorescently tagged Smt3 in the nucleolus when deconjugation is impaired (149). In conditional triple mutants lacking the three E3 SUMO ligases (Siz1, Siz2 and Mms21), rDNA stability is severely impaired (150). Several SUMOylation targets responsible for the observed rDNA instability have been studied. Top1 and Top2 are examples of proteins that are SUMOylated by Siz1 and Siz2, and contribute to rDNA stability by facilitating rDNA replication and transcription (150). Both top2ΔC (completely defective in SUMOylation), and top2-SNM mutants (partially defective in SUMOylation) exhibit a decrease in rDNA number (150). These mutants show altered localization at the rDNA locus, and when combined with Δtop1 mms21-CH result in synthetic lethality (150). This suggests redundancy of SUMO-Top2 with Top1 and some Mms21 SUMOylation substrates in maintenance of rDNA locus integrity. Top1 also gets SUMOylated and its SUMOylation contributes to rDNA stability, as seen by the rDNA stability defects in cells expressing non-SUMOylatable Top1 (Top1KR3) (150). In addition to Smc5, subunits of cohesin and condensin (Smc1, 2 and 3) were identified as Mms21 SUMOylation substrates (150). The SUMOylation of cohesin and condensin subunits is essential for rDNA maintenance and their binding to 5S rDNA region (150). The Smc5/6 complex was shown to be important for chromosome segregation at repetitive sequences. smc5 and smc6 conditional mutants display accumulation of X-shaped structures at the rDNA region as well as Rad52-dependent hyper-recombination (151,152). In addition to Slx5-Slx8, the Smc5/6 complex also contributes to the exclusion of Rad52 foci from the nucleolus, thereby protecting these regions from repeat expansion or contraction during recombination (152). Interestingly, a DSB induced in rDNA requires its transient exit from the nucleolus to be repaired by the nuclear Rad52 pool [(152), Figure 4D].
Other SUMOylated substrates that were recently identified are the nucleolus associated proteins Net1 and Fob1, as well as Tof2 (153), which also shows increased SUMOylation upon MMS-induced DNA damage (36). Net1 is part of the RENT complex, and together with Tof2, play a role in silencing the rDNA region, inhibiting recombination and repressing RNA Pol II transcription (154). Fob1 acts to block the progression of the replication fork and recruits subunits of the RENT complex and Tof2 (154,155). These proteins were found to be hyper-SUMOylated in Δulp2, Δslx5 and Δslx5Δulp2 mutants with reduced binding to rDNA in Δulp2 mutants that is rescued in absence of Slx5 (153). This suggests that the hyper-SUMOylation that occurs when deconjugation is impaired, targets them for Slx5-Slx8-mediated ubiquitylation and possibly proteasomal degradation (153). Hyper-SUMOylated-ubiquitylated Net1, Fob1 and Tof2 could be the more deleterious species that cannot bind rDNA resulting in hyper-recombination at these regions (153). The defects of modified Net1, Fob1 and Tof2 in rDNA binding, partly explains the rDNA defects observed in Δulp2 mutants. It remains, however, to identify the role of the basal level of SUMOylation of these proteins in rDNA maintenance and the DDR.
Telomeres
Telomeres represent another specialized DNA structure that requires dedicated machinery to protect and replicate (148). Telomeres are the ends of the linear DNA molecule that forms the chromosome. They consist of 75–150 repeats of C1-3A/TG1-3 with a terminal 3΄-tail called the G-tail followed by sub-telomeric regions called the X and Y’ regions, which also consist of repetitive sequences (156,157). Telomeres resemble DSBs and thus have to be carefully distinguished from them to avoid recombinational or end-joining repair (158). They pose a particular challenge for replication by the replication machinery and are therefore subject to shortening and erosion with each round of cell division. This necessitates a special DNA polymerase that belongs to the family of reverse transcriptases to replicate it called telomerase. The telomerase complex consists of several subunits, Est1, Est2 (catalytic subunit), Est3 and TLC1 (telomerase RNA) (156). Special proteins bind to the telomeric DNA to make up the telomere. Cdc13 binds the ssDNA at the G-tails and together with Stn1 and Ten1 form a complex resembling RPA. Rap1 protein binds the double-stranded TG repeat region, and together with its interacting partners Rif1 and Rif2, inhibit the telomerase activator Tel1 (156,157). The Yku70-Yku80 complex binds telomeric ends, similar to how it functions at DSBs, and protects DNA ends from resection by nucleases, whereas the Sir2-Sir3-Sir4 complex functions to silence telomeric regions. The Yku70-Yku80 complex and the Sir2-Sir3-Sir4 complex also tether telomeric ends to the inner nuclear membrane through interaction with the inner nuclear membrane protein Esc2 [(159–161) and Figure 4C]. This anchoring, however, is dynamic and subject to regulation by post-translational modifications (162). HR provides an alternative way of lengthening critically short telomeres in telomerase-deficient cells (163). Additional information on telomeres can be found in other reviews (156,158,164).
The high SUMOylation status of several telomeric proteins such as Yku70-Yku80, Sir4 and Esc2, indicates the importance of the SUMO signal in telomere maintenance. While single mutants of each of the three E3 ligases (Siz1, Siz2 and Mms21) exhibit longer telomeres (165), only Δsiz2 mutants exhibit loss of telomere anchoring (166). The longer telomeres seen in Δsiz2 mutants were shown to be due to telomerase mediated extension, not Rad52-dependent recombinational lengthening and was epistatic to Δpif1, which is a helicase that inhibits telomerase by displacing it at telomeres (166). On the other hand, the loss of telomere anchoring was not due to the disruption of Sir-dependent silencing or complex formation, but rather due to decreased interaction of both the Yku70-Yku80 complex and the Sir complex with Esc2 (166). Siz2 was also found to be the major E3 SUMOylating Sir4 and Yku80, as well as contribute to the SUMOylation of Yku70. Together, these findings suggest that the Siz2-dependent SUMOylation of the Yku70-Yku80 and Sir-complexes, and possibly other targets, promotes anchoring of telomeres to the nuclear envelope and inhibits telomerase through a pathway involving the Pif1 helicase (166).
The Smc5/6 complex, which includes Mms21, is constitutively found at telomeres (167). Multiple growth defects resulting from increased senescence were observed in mutants of the Smc5/6 complex (168,169). These result from shorter telomeres as observed in nse3-1 mutants (167), and defects in telomere clustering as observed in mms21-11 mutants (170). This highlights the role of Smc5/6 in telomere maintenance and its efficient replication (Figure 4C). In telomerase deficient cells, Smc5/6 helps resolve intermediates of HR mediated alternative lengthening of telomeres, thus slowing senescence (171). Upon MMS damage, the Smc5/6 complex shows enhanced enrichment at sub-telomeric regions, in a manner depending on the Mms21 subunit (172). It is unclear, however, whether it is strictly the SUMO ligase activity of Smc5/6 complex or the structural maintenance activity of the complex that may be affected by a defect in SUMO ligation, or both, that is required for its function at the telomeres. Either way, these findings indicate that SUMO and the Smc5/6 complex contribute to tethering telomeres to the nuclear envelope and resolving intermediates that arise during replication and recombination at the telomeres.
In addition to the role of SUMO in maintaining telomere anchoring, it also affects the stability and activity of the telomere-associated proteins upon DNA damage. MMS treatment induces the SUMOylation of several telomere binding proteins, such as Rap1, Cdc13, Pif1 and Yku70-Yku80 (165). The effects of their SUMOylation, if known, are listed in Table 1.
Uls1 has also been implicated in maintaining telomere end-joining inhibition through its ubiquitin ligase activity. While its ubiquitin ligase activity was never demonstrated in vitro, there is strong reason to assume it (118). Uls1 possesses a RING finger domain, multiple SIMs and was shown to regulate the levels of poly-SUMOylated Rap1, all typical of a STUbL (173). Rap1 is a telomere binding protein that inhibits NHEJ, protects the telomeric ends from nuclease activity and has a role in checkpoint signaling. Rap1 was shown to be SUMOylated at K240 and K246, resulting in decreased NHEJ inhibition activity except through the pathway that depends on Sir4 (173). Poly-SUMOylated Rap1 accumulates in Δuls1 mutants, and telomere fusions are observed in uls1 null, translocase and E3 ligase mutants (173). These telomere fusion events were shown to depend on SUMOylatable Rap1 and poly-SUMOylatable Smt3 (173). Together, this suggests that Uls1 mediates the proteasomal degradation of poly-SUMOylated Rap1, clearing the non-functional forms and allowing for the unmodified functional Rap1 molecules to bind, ensuring permanent NHEJ inhibition (173). The role of Uls1 in clearing SUMOylated Rap1 from telomeres together with its function in dislodging Rad51 nucleofilaments, suggest a role for Uls1 as a general molecular sweeper to dislodge proteins through its translocase and/or ubiquitin ligase activities (173).
In contrast to the activities of SUMO in inhibiting recombination at telomeres as discussed above, SUMOylation of Sgs1 has been shown to promote telomere–telomere recombination (174) and thus provides a means for alternative lengthening of telomeres in telomerase-deficient cells. Mutants expressing non-SUMOylatable Sgs1 (sgs1-K621R mutants) exhibited less telomere–telomere recombination in telomerase-deficient cells particularly in the formation of Type II recombinants, which show amplified telomeric repeats (174). On the other hand, the SUMOylation of Sgs1 was shown not to be important in recombinational repair, replication intermediates resolution, or rDNA recombination (174). The rescue of short telomeres in telomerase-deficient cells by recombination indicates their recognition as DSBs (175,176). This also requires the relocalization of the telomeres from the nuclear envelope to the nuclear pores, which contain several of the SUMO pathway proteins, indicating a regulatory role for SUMO in recombinational repair at telomeres (135).
SUMO AND STUBLS IN THE MAINTENANCE OF THE 2 μm CIRCLE
The 2 μm circle is an endogenous 6.4 kb plasmid that is normally found in almost all S. cerevisiae strains (177). It's a harmless parasitic plasmid that got acquired into the yeast nucleus, and is maintained at around 60 copies per cell (178). While the 2 μm circle provides no growth advantages, its overamplification was shown to cause multiple growth defects, such as cold sensitivity, irregularly shaped (nibbled) colonies and G2/M arrest, and these defects are often referred to as clonal lethality (25). The accumulation of high molecular weight aggregates of the plasmid underlies these growth defects that are observed in most of the SUMO pathway mutants (26,120,179). A mutation in the Ulp1 SUMO deconjugating enzyme was the first to be reported as causing the nibbled appearance, and this allele was initially named nib1 (25,26).
The 2 μm circle encodes four proteins, Rep1, Rep2, Raf1 and Flp1. Rep1 and Rep2 proteins are responsible for the proper segregation of the 2 μm circle to the daughter cells, through the interaction with the cis-acting DNA element, STB or REP3. The association of Rep1 and Rep2 with STB recruits the kinesin related motor Kip1, which exchanges the canonical H3 with Cse4, thus allowing STB to be recognized as a centromere (180,181). The proper propagation also involves the Rsc2 subunit of the RSC chromatin remodelling complex, as well as cohesin (182,183). Flp1 is a recombinase that is responsible for maintaining the high copy number of the 2 μm circle. Flp1 expression is tightly controlled in response to the levels of the 2μm circle. Rep1, Rep2 and Flp1 are SUMOylated and this SUMOylation is essential for maintaining the 2 μm circle levels (120,179,184). Mutants expressing non-SUMOylatable Flp1 exhibit clonal lethality, in a manner dependent on the HR pathway (179). Presumably, non-SUMOylated Flp1 induces DNA damage that gets repaired by the BIR subpathway of HR, resulting in hyperamplification and accumulation of abberant species of the 2 μm circle (179). An attractive model is that SUMOylated Flp1 levels are regulated by Slx5-Slx8 mediated degradation, and this would explain the clonal lethality observed in Δslx5 and Δslx8 mutants, that is dependent on HR proteins (106). The accumulation of Flp1 in SUMOylated or ubiquitylated forms in Δslx5 and Δslx8 mutants, however, has not yet been observed. The SUMOylation of Rep1 and Rep2 also contributes to the proper propagation of the plasmid by allowing the proper association with STB (184). This closely resembles the SUMO-dependent targeting of proteins, such as Top2 and the kinetochore proteins, to the centromere (185,186). The maintenance of the 2 μm circle provides an additional example in which SUMO participates in genome integrity and inheritance.
CONCLUSION
Since the discovery of ubiquitin more than three decades ago, our understanding of how complex cellular processes are regulated has significantly increased. Post-translational modifications using small chemical groups had already been appreciated as molecular regulators of cellular processes and signaling, and the further extension into ubiquitylation and SUMOylation has advanced our understanding further. The importance of ubiquitin and SUMO is obvious by the inviability of cells lacking them or the main enzymes involved in their processing. They were shown to act as true molecular switches to activate and regulate diverse pathways including the highly complicated DDR. They act through modifying interactions of repair proteins with other proteins or DNA, thus affecting their activities and the choice of repair pathway. The high reversibility of the process by deconjugating enzymes allows rapid and dynamic responses to cellular requirements. The high conservation of ubiquitin and SUMO in eukaryotic cells, as well as their processing enzymes, illustrate their importance as regulatory mechanisms among all species. Studying the DDR and how the cell repairs DNA damage increases our understanding of how mutagenesis and carcinogenesis occur and shed light on the possible mechanisms for cure. Due to the high conservation in the DNA repair pathways, most of the studies that have advanced our understanding on the roles of ubiquitin and SUMO in DNA repair have been conducted in yeast, however, still much of the process remains a mystery.
While the ubiquitin role in DNA repair is increasingly being shown to depend on altering interactions and signaling, still much of its role depends on the proteasome. This is evident by the presence of the 26S proteasomal complex coupled with DNA repair lesions where it mediates disassembly of factors to clear the way for the efficient assembly of repair and checkpoint machinery. A common characteristic to both ubiquitin and SUMO is the high specificity in the signal receptors. The residue modified, chain length and chain topology all affect signal transduction and outcome in a highly specific manner. The length of the SUMO chain in particular has been shown to be important for the choice of the perinuclear localization site for persistent DSBs.
Perhaps the most prominent role of SUMO is the regulation of HR. As much as HR is crucial for cell survival under DNA damaging conditions, its activity needs to be tightly regulated to prevent unnecessary recombination particularly during replication at repeat regions, which could cause gene amplifications and deletions contributing to carcinogenesis. The SUMOylation wave that is activated upon DNA damage affects the bulk of repair machinery in a manner dependent on proximity to the damage site. This spatial modification allows for a very small amount of modified species to exert the desired action due to high local concentration of the modification, while the majority of the protein remains unmodified. Indeed, while the non-specific nature of SUMO conjugation at first instance would seem like a random event, it leads to a very precise outcome. At the general level, SUMO modifications in response to DNA damage seems to function as a ‘molecular glue’, to facilitate the assembly of repair complexes and their recruitment owing to presence of high amount of SIMs on the repair factors. The disassembly of these repair foci also seems to be regulated by SUMOylation, partly through the action of STUbLs that target modified proteins to proteasomal degradation.
Proper understanding of the SUMOylation wave and its implications requires detailed study of each modified protein and the effect of SUMOylation on its activity and interactions. Particular challenges are the very low amount of SUMOylated proteins at any given time, and the high reversibility of the SUMO signal. A large amount of the protein does not need to be SUMOylated since the effects of the SUMO modifications are localized. Another factor that complicates the understanding of the SUMO pathway, is the promiscuity of the SUMO enzymes, which makes it hard to decipher the effects of the deficiency of the SUMO signal on a particular repair pathway. The experimental techniques employed pose a challenge in themselves. Most of them utilize SUMO deficient mutants to study a particular facet of the repair process. This, however, is biased to the repair assay used and disregards the complete overall effect of the SUMOylation wave that works on multiple parallel, and sometimes redundant, levels. Identifying the SUMOylated residues to make the SUMO deficient mutants is also very challenging. In several cases multiple lysines are targets for SUMOylation, and mutating one results in an enhanced SUMOylation of another, that would not normally be modified. Additionally, in numerous cases, more than one SUMO E3 ligases can SUMOylate a protein, raising questions on the specificity of the whole process, and how the signal is properly delivered. Thus, more studies are required to properly understand the intricacies of the SUMO pathway and the diversity of the roles it plays.
ACKNOWLEDGEMENTS
The authors thank Zeina Al-Natour for valuable comments, suggestions and critical reading of the article.
FUNDING
Funding for open access charge: United Arab Emirates University-UPAR; Terry Fox Foundation [to A.H.H].
Conflict of interest statement. The authors declare that there are no conflicts of interest.
REFERENCES
- 1.Kirkin V., Dikic I.. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr. Opin. Cell Biol. 2007; 19:199–205. [DOI] [PubMed] [Google Scholar]
- 2.van der Veen A.G., Ploegh H.L.. Ubiquitin-like proteins. Annu. Rev. Biochem. 2012; 81:323–357. [DOI] [PubMed] [Google Scholar]
- 3.Komander D., Rape M.. The ubiquitin code. Annu. Rev. Biochem. 2012; 81:203–229. [DOI] [PubMed] [Google Scholar]
- 4.Pickart C.M. Ubiquitin in chains. Trends Biochem. Sci. 2000; 25:544–548. [DOI] [PubMed] [Google Scholar]
- 5.Finley D., Ulrich H.D., Sommer T., Kaiser P.. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics. 2012; 192:319–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hay R.T. Decoding the SUMO signal. Biochem. Soc. Trans. 2013; 41:463–473. [DOI] [PubMed] [Google Scholar]
- 7.Praefcke G.J., Hofmann K., Dohmen R.J.. SUMO playing tag with ubiquitin. Trends Biochem. Sci. 2012; 37:23–31. [DOI] [PubMed] [Google Scholar]
- 8.Muller S., Hoege C., Pyrowolakis G., Jentsch S.. SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol. 2001; 2:202–210. [DOI] [PubMed] [Google Scholar]
- 9.Bartek J., Lukas J.. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 2007; 19:238–245. [DOI] [PubMed] [Google Scholar]
- 10.Boiteux S., Jinks-Robertson S.. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics. 2013; 193:1025–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulrich H.D. The SUMO system: an overview. Methods Mol. Biol. 2009; 497:3–16. [DOI] [PubMed] [Google Scholar]
- 12.Hay R.T. SUMO: a history of modification. Mol. Cell. 2005; 18:1–12. [DOI] [PubMed] [Google Scholar]
- 13.Johnson E.S., Schwienhorst I., Dohmen R.J., Blobel G.. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997; 16:5509–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindroos H.B., Strom L., Itoh T., Katou Y., Shirahige K., Sjogren C.. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell. 2006; 22:755–767. [DOI] [PubMed] [Google Scholar]
- 15.Johnson E.S., Gupta A.A.. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001; 106:735–744. [DOI] [PubMed] [Google Scholar]
- 16.Zhou W., Ryan J.J., Zhou H.. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J. Biol. Chem. 2004; 279:32262–32268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S.. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002; 419:135–141. [DOI] [PubMed] [Google Scholar]
- 18.Mullen J.R., Brill S.J.. Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J. Biol. Chem. 2008; 283:19912–19921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bylebyl G.R., Belichenko I., Johnson E.S.. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 2003; 278:44113–44120. [DOI] [PubMed] [Google Scholar]
- 20.Palancade B., Liu X., Garcia-Rubio M., Aguilera A., Zhao X., Doye V.. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol. Biol. Cell. 2007; 18:2912–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elmore Z.C., Donaher M., Matson B.C., Murphy H., Westerbeck J.W., Kerscher O.. Sumo-dependent substrate targeting of the SUMO protease Ulp1. BMC Biol. 2011; 9:74–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panse V.G., Kuster B., Gerstberger T., Hurt E.. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat. Cell Biol. 2003; 5:21–27. [DOI] [PubMed] [Google Scholar]
- 23.Hickey C.M., Wilson N.R., Hochstrasser M.. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 2012; 13:755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S.J., Hochstrasser M.. A new protease required for cell-cycle progression in yeast. Nature. 1999; 398:246–251. [DOI] [PubMed] [Google Scholar]
- 25.Holm C. Clonal lethality caused by the yeast plasmid 2 mu DNA. Cell. 1982; 29:585–594. [DOI] [PubMed] [Google Scholar]
- 26.Dobson M.J., Pickett A.J., Velmurugan S., Pinder J.B., Barrett L.A., Jayaram M., Chew J.S.. The 2 microm plasmid causes cell death in Saccharomyces cerevisiae with a mutation in Ulp1 protease. Mol. Cell. Biol. 2005; 25:4299–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckhoff J., Dohmen R.J.. In vitro studies reveal a sequential mode of chain processing by the yeast SUMO (Small Ubiquitin-related Modifier)-specific protease Ulp2. J. Biol. Chem. 2015; 290:12268–12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S.J., Hochstrasser M.. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 2000; 20:2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Danielsen J.R., Povlsen L.K., Villumsen B.H., Streicher W., Nilsson J., Wikstrom M., Bekker-Jensen S., Mailand N.. DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J. Cell Biol. 2012; 197:179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song J., Durrin L.K., Wilkinson T.A., Krontiris T.G., Chen Y.. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:14373–14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatham M.H., Geoffroy M.C., Shen L., Plechanovova A., Hattersley N., Jaffray E.G. et al. . RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008; 10:538–546. [DOI] [PubMed] [Google Scholar]
- 32.Bologna S., Altmannova V., Valtorta E., Koenig C., Liberali P., Gentili C. et al. . Sumoylation regulates EXO1 stability and processing of DNA damage. Cell Cycle. 2015; 14:2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Branzei D., Vanoli F., Foiani M.. SUMOylation regulates Rad18-mediated template switch. Nature. 2008; 456:915–920. [DOI] [PubMed] [Google Scholar]
- 34.Kolesar P., Altmannova V., Silva S., Lisby M., Krejci L.. Pro-recombination role of Srs2 requires SUMO but is independent of PCNA interaction. J. Biol. Chem. 2016; 291:7594–7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bermudez-Lopez M., Pocino-Merino I., Sanchez H., Bueno A., Guasch C., Almedawar S., Bru-Virgili S., Garí E., Wyman C., Reverter D. et al. . ATPase-dependent control of the Mms21 SUMO ligase during DNA repair. PLoS Biol. 2015; 13:e1002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cremona C.A., Sarangi P., Yang Y., Hang L.E., Rahman S., Zhao X.. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol. Cell. 2012; 45:422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Psakhye I., Jentsch S.. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012; 151:807–820. [DOI] [PubMed] [Google Scholar]
- 38.Sriramachandran A.M., Dohmen R.J.. SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta. 2014; 1843:75–85. [DOI] [PubMed] [Google Scholar]
- 39.Rape M., Hoppe T., Gorr I., Kalocay M., Richly H., Jentsch S.. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001; 107:667–677. [DOI] [PubMed] [Google Scholar]
- 40.Shcherbik N., Haines D.S.. Cdc48p(Npl4p/Ufd1p) binds and segregates membrane-anchored/tethered complexes via a polyubiquitin signal present on the anchors. Mol. Cell. 2007; 25:385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bauer N.C., Corbett A.H., Doetsch P.W.. The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res. 2015; 43:10083–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffiths L.M., Swartzlander D., Meadows K.L., Wilkinson K.D., Corbett A.H., Doetsch P.W.. Dynamic compartmentalization of base excision repair proteins in response to nuclear and mitochondrial oxidative stress. Mol. Cell. Biol. 2009; 29:794–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alseth I., Eide L., Pirovano M., Rognes T., Seeberg E., Bjoras M.. The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell. Biol. 1999; 19:3779–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.You H.J., Swanson R.L., Harrington C., Corbett A.H., Jinks-Robertson S., Senturker S., Wallace S.S., Boiteux S., Dizdaroglu M., Doetsch P.W.. Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry. 1999; 38:11298–11306. [DOI] [PubMed] [Google Scholar]
- 45.Li S. Transcription coupled nucleotide excision repair in the yeast Saccharomyces cerevisiae: the ambiguous role of Rad26. DNA Repair (Amst). 2015; 36:43–48. [DOI] [PubMed] [Google Scholar]
- 46.Silver H.R., Nissley J.A., Reed S.H., Hou Y.M., Johnson E.S.. A role for SUMO in nucleotide excision repair. DNA Repair (Amst). 2011; 10:1243–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarangi P., Bartosova Z., Altmannova V., Holland C., Chavdarova M., Lee S.E., Krejci L., Zhao X.. Sumoylation of the Rad1 nuclease promotes DNA repair and regulates its DNA association. Nucleic Acids Res. 2014; 42:6393–6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toh G.W., Sugawara N., Dong J., Toth R., Lee S.E., Haber J.E., Rouse J.. Mec1/Tel1-dependent phosphorylation of Slx4 stimulates Rad1-Rad10-dependent cleavage of non-homologous DNA tails. DNA Repair (Amst). 2010; 9:718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolesar P., Sarangi P., Altmannova V., Zhao X., Krejci L.. Dual roles of the SUMO-interacting motif in the regulation of Srs2 sumoylation. Nucleic Acids Res. 2012; 40:7831–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stingele J., Schwarz M.S., Bloemeke N., Wolf P.G., Jentsch S.. A DNA-dependent protease involved in DNA-protein crosslink repair. Cell. 2014; 158:327–338. [DOI] [PubMed] [Google Scholar]
- 51.Pommier Y., Barcelo J.M., Rao V.A., Sordet O., Jobson A.G., Thibaut L. et al. . Repair of topoisomerase I-mediated DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 2006; 81:179–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iyer L.M., Koonin E.V., Aravind L.. Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle. 2004; 3:1440–1450. [DOI] [PubMed] [Google Scholar]
- 53.Balakirev M.Y., Mullally J.E., Favier A., Assard N., Sulpice E., Lindsey D.F., Rulina A.V., Gidrol X., Wilkinson K.D.. Wss1 metalloprotease partners with Cdc48/Doa1 in processing genotoxic SUMO conjugates. eLife. 2015; 4:e06763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prakash S., Sung P., Prakash L.. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet. 1993; 27:33–70. [DOI] [PubMed] [Google Scholar]
- 55.Ulrich H.D. Degradation or maintenance: actions of the ubiquitin system on eukaryotic chromatin. Eukaryot. Cell. 2002; 1:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsutakawa S.E., Yan C., Xu X., Weinacht C.P., Freudenthal B.D., Yang K., Zhuang Z., Washington M.T., Tainer J.A., Ivanov I.. Structurally distinct ubiquitin- and sumo-modified PCNA: implications for their distinct roles in the DNA damage response. Structure. 2015; 23:724–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee K.Y., Myung K.. PCNA modifications for regulation of post-replication repair pathways. Mol. Cells. 2008; 26:5–11. [PMC free article] [PubMed] [Google Scholar]
- 58.Strzalka W., Ziemienowicz A.. Proliferating cell nuclear antigen (PCNA): a key factor in DNA replication and cell cycle regulation. Ann. Bot. 2011; 107:1127–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu Q., Chang Y., Yang J., Wei Q.. Post-translational modifications of proliferating cell nuclear antigen: a key signal integrator for DNA damage response (Review). Oncol. Lett. 2014; 7:1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Torres-Ramos C.A., Prakash S., Prakash L.. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 2002; 22:2419–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao W., Chow B.L., Fontanie T., Ma L., Bacchetti S., Hryciw T., Broomfield S.. Genetic interactions between error-prone and error-free postreplication repair pathways in Saccharomyces cerevisiae. Mutat. Res. 1999; 435:1–11. [DOI] [PubMed] [Google Scholar]
- 62.Xiao W., Chow B.L., Broomfield S., Hanna M.. The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics. 2000; 155:1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waters L.S., Minesinger B.K., Wiltrout M.E., D'Souza S., Woodruff R.V., Walker G.C.. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 2009; 73:134–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blastyak A., Pinter L., Unk I., Prakash L., Prakash S., Haracska L.. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell. 2007; 28:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi K., Batke S., Szakal B., Lowther J., Hao F., Sarangi P., Branzei D., Ulrich H.D., Zhao X.. Concerted and differential actions of two enzymatic domains underlie Rad5 contributions to DNA damage tolerance. Nucleic Acids Res. 2015; 43:2666–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Branzei D. Ubiquitin family modifications and template switching. FEBS Lett. 2011; 585:2810–2817. [DOI] [PubMed] [Google Scholar]
- 67.Bi X. Mechanism of DNA damage tolerance. World J. Biol. Chem. 2015; 6:48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prado F. Homologous recombination maintenance of genome integrity during DNA damage tolerance. Mol. Cell. Oncol. 2014; 1:e957039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karras G.I., Jentsch S.. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010; 141:255–267. [DOI] [PubMed] [Google Scholar]
- 70.Pfander B., Moldovan G.L., Sacher M., Hoege C., Jentsch S.. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005; 436:428–433. [DOI] [PubMed] [Google Scholar]
- 71.Parker J.L., Bucceri A., Davies A.A., Heidrich K., Windecker H., Ulrich H.D.. SUMO modification of PCNA is controlled by DNA. EMBO J. 2008; 27:2422–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Papouli E., Chen S., Davies A.A., Huttner D., Krejci L., Sung P., Ulrich H.D.. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005; 19:123–133. [DOI] [PubMed] [Google Scholar]
- 73.Windecker H., Ulrich H.D.. Architecture and assembly of poly-SUMO chains on PCNA in Saccharomyces cerevisiae. J. Mol. Biol. 2008; 376:221–231. [DOI] [PubMed] [Google Scholar]
- 74.Parnas O., Zipin-Roitman A., Pfander B., Liefshitz B., Mazor Y., Ben-Aroya S., Jentsch S., Kupiec M.. Elg1, an alternative subunit of the RFC clamp loader, preferentially interacts with SUMOylated PCNA. EMBO J. 2010; 29:2611–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kubota T., Nishimura K., Kanemaki M.T., Donaldson A.D.. The Elg1 replication factor C-like complex functions in PCNA unloading during DNA replication. Mol. Cell. 2013; 50:273–280. [DOI] [PubMed] [Google Scholar]
- 76.Das-Bradoo S., Nguyen H.D., Wood J.L., Ricke R.M., Haworth J.C., Bielinsky A.K.. Defects in DNA ligase I trigger PCNA ubiquitylation at Lys 107. Nat. Cell Biol. 2010; 12:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moldovan G.L., Pfander B., Jentsch S.. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell. 2006; 23:723–732. [DOI] [PubMed] [Google Scholar]
- 78.Parker J.L., Ulrich H.D.. A SUMO-interacting motif activates budding yeast ubiquitin ligase Rad18 towards SUMO-modified PCNA. Nucleic Acids Res. 2012; 40:11380–11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., Seki M., Enomoto T., Ohta K., Foiani M.. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006; 127:509–522. [DOI] [PubMed] [Google Scholar]
- 80.Parker J.L., Ulrich H.D.. SIM-dependent enhancement of substrate-specific SUMOylation by a ubiquitin ligase in vitro. Biochem. J. 2014; 457:435–440. [DOI] [PubMed] [Google Scholar]
- 81.Ulrich H.D. The srs2 suppressor of UV sensitivity acts specifically on the RAD5- and MMS2-dependent branch of the RAD6 pathway. Nucleic Acids Res. 2001; 29:3487–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Broomfield S., Xiao W.. Suppression of genetic defects within the RAD6 pathway by srs2 is specific for error-free post-replication repair but not for damage-induced mutagenesis. Nucleic Acids Res. 2002; 30:732–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Antony E., Tomko E.J., Xiao Q., Krejci L., Lohman T.M., Ellenberger T.. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell. 2009; 35:105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Le Breton C., Dupaigne P., Robert T., Le Cam E., Gangloff S., Fabre F., Veaute X.. Srs2 removes deadly recombination intermediates independently of its interaction with SUMO-modified PCNA. Nucleic Acids Res. 2008; 36:4964–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Burkovics P., Sebesta M., Sisakova A., Plault N., Szukacsov V., Robert T., Pinter L., Marini V., Kolesar P., Haracska L. et al. . Srs2 mediates PCNA-SUMO-dependent inhibition of DNA repair synthesis. EMBO J. 2013; 32:742–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Halas A., Krawczyk M., Sledziewska-Gojska E.. PCNA SUMOylation protects against PCNA polyubiquitination-mediated, Rad59-dependent, spontaneous, intrachromosomal gene conversion. Mutat. Res. 2016; 791-792:10–18. [DOI] [PubMed] [Google Scholar]
- 87.Bermudez-Lopez M., Ceschia A., de Piccoli G., Colomina N., Pasero P., Aragon L., Torres-Rosell J.. The Smc5/6 complex is required for dissolution of DNA-mediated sister chromatid linkages. Nucleic Acids Res. 2010; 38:6502–6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stephan A.K., Kliszczak M., Morrison C.G.. The Nse2/Mms21 SUMO ligase of the Smc5/6 complex in the maintenance of genome stability. FEBS Lett. 2011; 585:2907–2913. [DOI] [PubMed] [Google Scholar]
- 89.Choi K., Szakal B., Chen Y.H., Branzei D., Zhao X.. The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol. Biol. Cell. 2010; 21:2306–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hang L.E., Peng J., Tan W., Szakal B., Menolfi D., Sheng Z., Lobachev K., Branzei D., Feng W., Zhao X.. Rtt107 is a multi-functional scaffold supporting replication progression with partner SUMO and ubiquitin ligases. Mol. Cell. 2015; 60:268–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wilson T.E., Grawunder U., Lieber M.R.. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997; 388:495–498. [DOI] [PubMed] [Google Scholar]
- 92.Krejci L., Altmannova V., Spirek M., Zhao X.. Homologous recombination and its regulation. Nucleic Acids Res. 2012; 40:5795–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Symington L.S., Rothstein R., Lisby M.. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014; 198:795–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mehta A., Haber J.E.. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014; 6:a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jasin M., Rothstein R.. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013; 5:a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Suzuki R., Shindo H., Tase A., Kikuchi Y., Shimizu M., Yamazaki T.. Solution structures and DNA binding properties of the N-terminal SAP domains of SUMO E3 ligases from Saccharomyces cerevisiae and Oryza sativa. Proteins. 2009; 75:336–347. [DOI] [PubMed] [Google Scholar]
- 97.Rytinki M.M., Kaikkonen S., Pehkonen P., Jaaskelainen T., Palvimo J.J.. PIAS proteins: pleiotropic interactors associated with SUMO. Cell. Mol. Life Sci. 2009; 66:3029–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chung I., Zhao X.. DNA break-induced sumoylation is enabled by collaboration between a SUMO ligase and the ssDNA-binding complex RPA. Genes Dev. 2015; 29:1593–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marechal A., Zou L.. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015; 25:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ohuchi T., Seki M., Branzei D., Maeda D., Ui A., Ogiwara H. et al. . Rad52 sumoylation and its involvement in the efficient induction of homologous recombination. DNA Repair (Amst). 2008; 7:879–889. [DOI] [PubMed] [Google Scholar]
- 101.Sacher M., Pfander B., Hoege C., Jentsch S.. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 2006; 8:1284–1290. [DOI] [PubMed] [Google Scholar]
- 102.Altmannova V., Eckert-Boulet N., Arneric M., Kolesar P., Chaloupkova R., Damborsky J., Sung P., Zhao X., Lisby M., Krejci L.. Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic Acids Res. 2010; 38:4708–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bergink S., Ammon T., Kern M., Schermelleh L., Leonhardt H., Jentsch S.. Role of Cdc48/p97 as a SUMO-targeted segregase curbing Rad51-Rad52 interaction. Nat. Cell Biol. 2013; 15:526–532. [DOI] [PubMed] [Google Scholar]
- 104.Esta A., Ma E., Dupaigne P., Maloisel L., Guerois R., Le Cam E., Veaute X., Coïc E.. Rad52 sumoylation prevents the toxicity of unproductive Rad51 filaments independently of the anti-recombinase Srs2. PLoS Genet. 2013; 9:e1003833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xie Y., Kerscher O., Kroetz M.B., McConchie H.F., Sung P., Hochstrasser M.. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 2007; 282:34176–34184. [DOI] [PubMed] [Google Scholar]
- 106.Burgess R.C., Rahman S., Lisby M., Rothstein R., Zhao X.. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol. Cell. Biol. 2007; 27:6153–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Su X.A., Dion V., Gasser S.M., Freudenreich C.H.. Regulation of recombination at yeast nuclear pores controls repair and triplet repeat stability. Genes Dev. 2015; 29:1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Friedl A.A., Liefshitz B., Steinlauf R., Kupiec M.. Deletion of the SRS2 gene suppresses elevated recombination and DNA damage sensitivity in rad5 and rad18 mutants of Saccharomyces cerevisiae. Mutat. Res. 2001; 486:137–146. [DOI] [PubMed] [Google Scholar]
- 109.Armstrong A.A., Mohideen F., Lima C.D.. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature. 2012; 483:59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Miura T., Shibata T., Kusano K.. Putative antirecombinase Srs2 DNA helicase promotes noncrossover homologous recombination avoiding loss of heterozygosity. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:16067–16072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hishida T., Hirade Y., Haruta N., Kubota Y., Iwasaki H.. Srs2 plays a critical role in reversible G2 arrest upon chronic and low doses of UV irradiation via two distinct homologous recombination-dependent mechanisms in postreplication repair-deficient cells. Mol. Cell. Biol. 2010; 30:4840–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ira G., Malkova A., Liberi G., Foiani M., Haber J.E.. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003; 115:401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sarangi P., Steinacher R., Altmannova V., Fu Q., Paull T.T., Krejci L., Whitby M.C., Zhao X.. Sumoylation influences DNA break repair partly by increasing the solubility of a conserved end resection protein. PLoS Genet. 2015; 11:e1004899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hang L.E., Lopez C.R., Liu X., Williams J.M., Chung I., Wei L., Bertuch A.A., Zhao X.. Regulation of Ku-DNA association by Yku70 C-terminal tail and SUMO modification. J. Biol. Chem. 2014; 289:10308–10317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vigasova D., Sarangi P., Kolesar P., Vlasakova D., Slezakova Z., Altmannova V., Nikulenkov F., Anrather D., Gith R., Zhao X. et al. . Lif1 SUMOylation and its role in non-homologous end-joining. Nucleic Acids Res. 2013; 41:5341–5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mullen J.R., Kaliraman V., Ibrahim S.S., Brill S.J.. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001; 157:103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ii T., Mullen J.R., Slagle C.E., Brill S.J.. Stimulation of in vitro sumoylation by Slx5-Slx8: evidence for a functional interaction with the SUMO pathway. DNA Repair (Amst). 2007; 6:1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Uzunova K., Gottsche K., Miteva M., Weisshaar S.R., Glanemann C., Schnellhardt M., Niessen M., Scheel H., Hofmann K., Johnson E.S. et al. . Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 2007; 282:34167–34175. [DOI] [PubMed] [Google Scholar]
- 119.Zhang C., Roberts T.M., Yang J., Desai R., Brown G.W.. Suppression of genomic instability by SLX5 and SLX8 in Saccharomyces cerevisiae. DNA Repair (Amst). 2006; 5:336–346. [DOI] [PubMed] [Google Scholar]
- 120.Chen X.L., Reindle A., Johnson E.S.. Misregulation of 2 microm circle copy number in a SUMO pathway mutant. Mol. Cell. Biol. 2005; 25:4311–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M.B., Roberts T.M., Brown G.W., Varela E., Hediger F., Gasser S.M., Krogan N.J.. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008; 322:597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bohm S., Mihalevic M.J., Casal M.A., Bernstein K.A.. Disruption of SUMO-targeted ubiquitin ligases Slx5-Slx8/RNF4 alters RecQ-like helicase Sgs1/BLM localization in yeast and human cells. DNA Repair (Amst). 2015; 26:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Oza P., Jaspersen S.L., Miele A., Dekker J., Peterson C.L.. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009; 23:912–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Oza P., Peterson C.L.. Opening the DNA repair toolbox: localization of DNA double strand breaks to the nuclear periphery. Cell Cycle. 2010; 9:43–49. [DOI] [PubMed] [Google Scholar]
- 125.Schober H., Ferreira H., Kalck V., Gehlen L.R., Gasser S.M.. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 2009; 23:928–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Texari L., Stutz F.. Sumoylation and transcription regulation at nuclear pores. Chromosoma. 2015; 124:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Akhtar A., Gasser S.M.. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 2007; 8:507–517. [DOI] [PubMed] [Google Scholar]
- 128.Mekhail K., Moazed D.. The nuclear envelope in genome organization, expression and stability. Nat. Rev. Mol. Cell Biol. 2010; 11:317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhao X., Wu C.Y., Blobel G.. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 2004; 167:605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li S.J., Hochstrasser M.. The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J. Cell Biol. 2003; 160:1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pennaneach V., Putnam C.D., Kolodner R.D.. Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae. Mol. Microbiol. 2006; 59:1357–1368. [DOI] [PubMed] [Google Scholar]
- 132.Kalocsay M., Hiller N.J., Jentsch S.. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell. 2009; 33:335–343. [DOI] [PubMed] [Google Scholar]
- 133.Gardner J.M., Smoyer C.J., Stensrud E.S., Alexander R., Gogol M., Wiegraebe W., Jaspersen S.L.. Targeting of the SUN protein Mps3 to the inner nuclear membrane by the histone variant H2A.Z. J. Cell Biol. 2011; 193:489–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gartenberg M.R. Life on the edge: telomeres and persistent DNA breaks converge at the nuclear periphery. Genes Dev. 2009; 23:1027–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Khadaroo B., Teixeira M.T., Luciano P., Eckert-Boulet N., Germann S.M., Simon M.N., I Gallina., Abdallah P., Gilson E., Géli V. et al. . The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat. Cell Biol. 2009; 11:980–987. [DOI] [PubMed] [Google Scholar]
- 136.Horigome C., Bustard D.E., Marcomini I., Delgoshaie N., Tsai-Pflugfelder M., Cobb J.A., Gasser S.M.. PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL. Genes Dev. 2016; 30:931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Alvaro D., Lisby M., Rothstein R.. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 2007; 3:e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Miura T., Yamana Y., Usui T., Ogawa H.I., Yamamoto M.T., Kusano K.. Homologous recombination via synthesis-dependent strand annealing in yeast requires the Irc20 and Srs2 DNA helicases. Genetics. 2012; 191:65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cal-Bakowska M., Litwin I., Bocer T., Wysocki R., Dziadkowiec D.. The Swi2-Snf2-like protein Uls1 is involved in replication stress response. Nucleic Acids Res. 2011; 39:8765–8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang Z., Buchman A.R.. Identification of a member of a DNA-dependent ATPase family that causes interference with silencing. Mol. Cell. Biol. 1997; 17:5461–5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tan W., Wang Z., Prelich G.. Physical and genetic interactions between Uls1 and the Slx5-Slx8 SUMO-targeted ubiquitin ligase. G3 (Bethesda). 2013; 3:771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shah P.P., Zheng X., Epshtein A., Carey J.N., Bishop D.K., Klein H.L.. Swi2/Snf2-related translocases prevent accumulation of toxic Rad51 complexes during mitotic growth. Mol. Cell. 2010; 39:862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kramarz K., Litwin I., Cal-Bakowska M., Szakal B., Branzei D., Wysocki R., Dziadkowiec D.. Swi2/Snf2-like protein Uls1 functions in the Sgs1-dependent pathway of maintenance of rDNA stability and alleviation of replication stress. DNA Repair (Amst). 2014; 21:24–35. [DOI] [PubMed] [Google Scholar]
- 144.Unk I., Hajdu I., Blastyak A., Haracska L.. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst). 2010; 9:257–267. [DOI] [PubMed] [Google Scholar]
- 145.Shiratori A., Shibata T., Arisawa M., Hanaoka F., Murakami Y., Eki T.. Systematic identification, classification, and characterization of the open reading frames which encode novel helicase-related proteins in Saccharomyces cerevisiae by gene disruption and Northern analysis. Yeast. 1999; 15:219–253. [DOI] [PubMed] [Google Scholar]
- 146.Richardson A., Gardner R.G., Prelich G.. Physical and genetic associations of the Irc20 ubiquitin ligase with Cdc48 and SUMO. PloS One. 2013; 8:e76424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jones G.M., Stalker J., Humphray S., West A., Cox T., Rogers J., Dunham I., Prelich G.. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat. Methods. 2008; 5:239–241. [DOI] [PubMed] [Google Scholar]
- 148.Marcomini I., Gasser S.M.. Nuclear organization in DNA end processing: Telomeres vs double-strand breaks. DNA Repair (Amst). 2015; 32:134–140. [DOI] [PubMed] [Google Scholar]
- 149.Takahashi Y., Strunnikov A.. In vivo modeling of polysumoylation uncovers targeting of Topoisomerase II to the nucleolus via optimal level of SUMO modification. Chromosoma. 2008; 117:189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Takahashi Y., Dulev S., Liu X., Hiller N.J., Zhao X., Strunnikov A.. Cooperation of sumoylated chromosomal proteins in rDNA maintenance. PLoS Genet. 2008; 4:e1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Torres-Rosell J., Machin F., Farmer S., Jarmuz A., Eydmann T., Dalgaard J.Z., Aragón L.. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat. Cell Biol. 2005; 7:412–419. [DOI] [PubMed] [Google Scholar]
- 152.Torres-Rosell J., Sunjevaric I., De Piccoli G., Sacher M., Eckert-Boulet N., Reid R., Jentsch S., Rothstein R., Aragón L., Lisby M.. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 2007; 9:923–931. [DOI] [PubMed] [Google Scholar]
- 153.Gillies J., Hickey C.M., Su D., Wu Z., Peng J., Hochstrasser M.. SUMO Pathway Modulation of Regulatory Protein Binding at the Ribosomal DNA Locus in Saccharomyces cerevisiae. Genetics. 2016; 202:1377–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Huang J., Brito I.L., Villen J., Gygi S.P., Amon A., Moazed D.. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006; 20:2887–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Waples W.G., Chahwan C., Ciechonska M., Lavoie B.D.. Putting the brake on FEAR: Tof2 promotes the biphasic release of Cdc14 phosphatase during mitotic exit. Mol. Biol. Cell. 2009; 20:245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wellinger R.J., Zakian V.A.. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics. 2012; 191:1073–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Grandin N., Charbonneau M.. Protection against chromosome degradation at the telomeres. Biochimie. 2008; 90:41–59. [DOI] [PubMed] [Google Scholar]
- 158.O'Sullivan R.J., Karlseder J.. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010; 11:171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Laroche T., Martin S.G., Gotta M., Gorham H.C., Pryde F.E., Louis E.J., Gasser S.M.. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 1998; 8:653–656. [DOI] [PubMed] [Google Scholar]
- 160.Mishra K., Shore D.. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr. Biol. 1999; 9:1123–1126. [DOI] [PubMed] [Google Scholar]
- 161.Taddei A., Hediger F., Neumann F.R., Bauer C., Gasser S.M.. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 2004; 23:1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Peuscher M.H., Jacobs J.J.. Posttranslational control of telomere maintenance and the telomere damage response. Cell Cycle. 2012; 11:1524–1534. [DOI] [PubMed] [Google Scholar]
- 163.Teng S.C., Chang J., McCowan B., Zakian V.A.. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell. 2000; 6:947–952. [DOI] [PubMed] [Google Scholar]
- 164.Malyavko A.N., Parfenova Y.Y., Zvereva M.I., Dontsova O.A.. Telomere length regulation in budding yeasts. FEBS Lett. 2014; 588:2530–2536. [DOI] [PubMed] [Google Scholar]
- 165.Hang L.E., Liu X., Cheung I., Yang Y., Zhao X.. SUMOylation regulates telomere length homeostasis by targeting Cdc13. Nat. Struct. Mol. Biol. 2011; 18:920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ferreira H.C., Luke B., Schober H., Kalck V., Lingner J., Gasser S.M.. The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat. Cell Biol. 2011; 13:867–874. [DOI] [PubMed] [Google Scholar]
- 167.Moradi-Fard S., Sarthi J., Tittel-Elmer M., Lalonde M., Cusanelli E., Chartrand P., Cobb J.A.. Smc5/6 is a telomere-associated complex that regulates Sir4 binding and TPE. PLoS Genet. 2016; 12:e1006268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Murray J.M., Carr A.M.. Smc5/6: a link between DNA repair and unidirectional replication. Nat. Rev. Mol. Cell Biol. 2008; 9:177–182. [DOI] [PubMed] [Google Scholar]
- 169.De Piccoli G., Torres-Rosell J., Aragon L.. The unnamed complex: what do we know about Smc5-Smc6. Chromosome Res. 2009; 17:251–263. [DOI] [PubMed] [Google Scholar]
- 170.Noel J.F., Wellinger R.J.. Abrupt telomere losses and reduced end-resection can explain accelerated senescence of Smc5/6 mutants lacking telomerase. DNA Repair (Amst). 2011; 10:271–282. [DOI] [PubMed] [Google Scholar]
- 171.Chavez A., George V., Agrawal V., Johnson F.B.. Sumoylation and the structural maintenance of chromosomes (Smc) 5/6 complex slow senescence through recombination intermediate resolution. J. Biol. Chem. 2010; 285:11922–11930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pebernard S., Schaffer L., Campbell D., Head S.R., Boddy M.N.. Localization of Smc5/6 to centromeres and telomeres requires heterochromatin and SUMO, respectively. EMBO J. 2008; 27:3011–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lescasse R., Pobiega S., Callebaut I., Marcand S.. End-joining inhibition at telomeres requires the translocase and polySUMO-dependent ubiquitin ligase Uls1. EMBO J. 2013; 32:805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lu C.Y., Tsai C.H., Brill S.J., Teng S.C.. Sumoylation of the BLM ortholog, Sgs1, promotes telomere-telomere recombination in budding yeast. Nucleic Acids Res. 2010; 38:488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lin Y.H., Chang C.C., Wong C.W., Teng S.C.. Recruitment of Rad51 and Rad52 to short telomeres triggers a Mec1-mediated hypersensitivity to double-stranded DNA breaks in senescent budding yeast. PloS One. 2009; 4:e8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Eckert-Boulet N., Lisby M.. Regulation of homologous recombination at telomeres in budding yeast. FEBS Lett. 2010; 584:3696–3702. [DOI] [PubMed] [Google Scholar]
- 177.Futcher A.B. The 2 micron circle plasmid of Saccharomyces cerevisiae. Yeast. 1988; 4:27–40. [DOI] [PubMed] [Google Scholar]
- 178.Falcon A.A., Rios N., Aris J.P.. 2-micron circle plasmids do not reduce yeast life span. FEMS Microbiol. Lett. 2005; 250:245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Xiong L., Chen X.L., Silver H.R., Ahmed N.T., Johnson E.S.. Deficient SUMO attachment to Flp recombinase leads to homologous recombination-dependent hyperamplification of the yeast 2 microm circle plasmid. Mol. Biol. Cell. 2009; 20:1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cui H., Ghosh S.K., Jayaram M.. The selfish yeast plasmid uses the nuclear motor Kip1p but not Cin8p for its localization and equal segregation. J. Cell Biol. 2009; 185:251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hajra S., Ghosh S.K., Jayaram M.. The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-microm circle partitioning locus and promotes equal plasmid segregation. J. Cell Biol. 2006; 174:779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mehta S., Yang X.M., Chan C.S., Dobson M.J., Jayaram M., Velmurugan S.. The 2 micron plasmid purloins the yeast cohesin complex: a mechanism for coupling plasmid partitioning and chromosome segregation. J. Cell Biol. 2002; 158:625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wong M.C., Scott-Drew S.R., Hayes M.J., Howard P.J., Murray J.A.. RSC2, encoding a component of the RSC nucleosome remodeling complex, is essential for 2 microm plasmid maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 2002; 22:4218–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pinder J.B., McQuaid M.E., Dobson M.J.. Deficient sumoylation of yeast 2-micron plasmid proteins Rep1 and Rep2 associated with their loss from the plasmid-partitioning locus and impaired plasmid inheritance. PLoS One. 2013; 8:e60384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Takahashi Y., Yong-Gonzalez V., Kikuchi Y., Strunnikov A.. SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics. 2006; 172:783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Montpetit B., Hazbun T.R., Fields S., Hieter P.. Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J. Cell Biol. 2006; 174:653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]