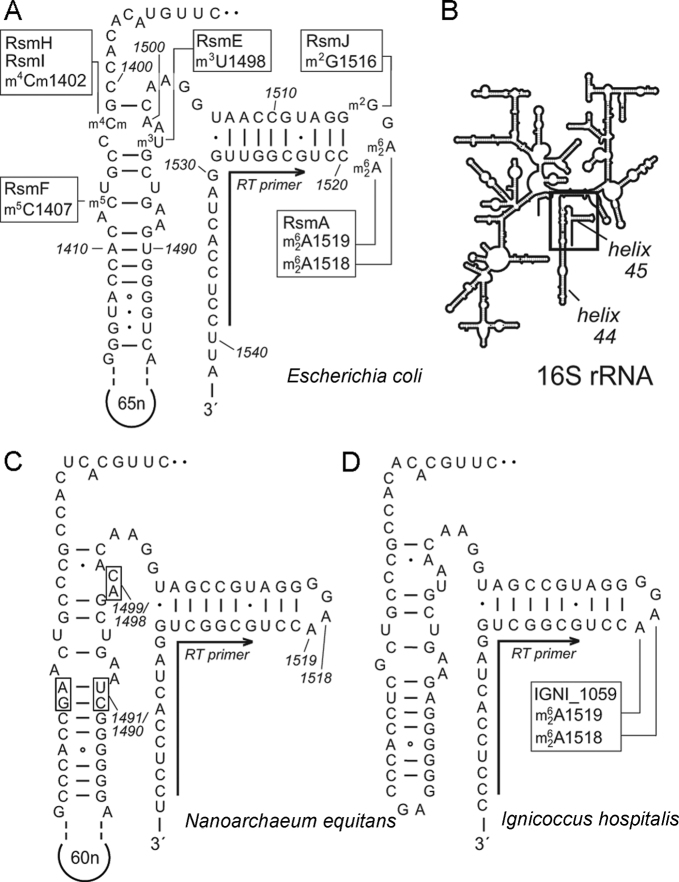
Figure 1.
(A) Secondary structures of helices 44 and 45 in 16S rRNA from Escherichia coli showing the sites of nucleotide methylation and the enzymes involved. These include RsmA, which N6, N6-dimethylates A1518 and A1519. All nucleotide annotations here use the E. coli numbering system. (B) The relative positions of helices 44 and 45 depicted on the 16S rRNA secondary structure schematic. (C) The corresponding structures in Nanoarchaeum equitans and (D) Ignicoccus hospitalis 16S rRNAs show a high degree of conservation in and around helix 45. Nucleotide variations that were used to distinguish and isolate the archaeal sequences are shown on the N. equitans structure (boxes and the 60 nucleotide extension in helix 44). A single oligonucleotide was used for reverse transcriptase (RT) primer extension reactions on all three rRNA, and hybridizes as indicated (arrows) to the conserved sequence at the 3΄-side of helix 45.