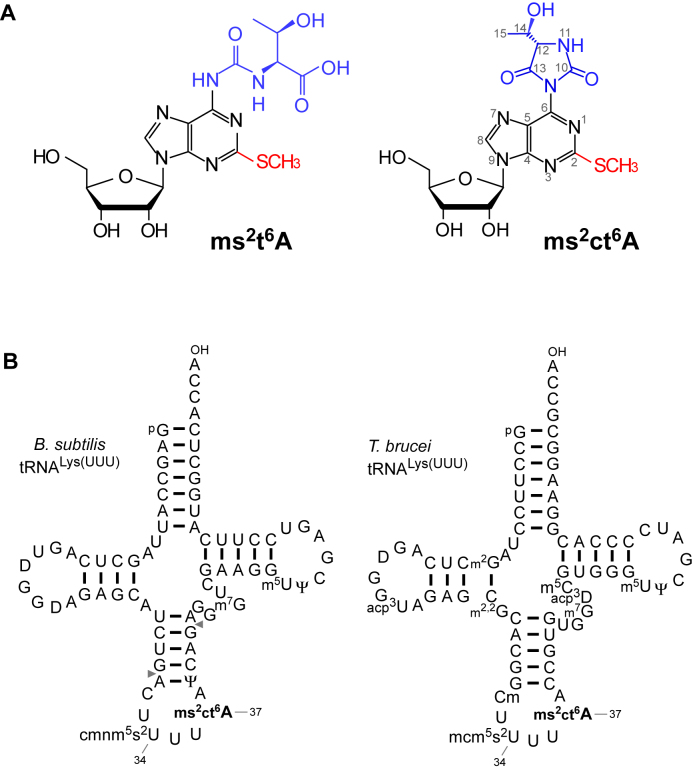
Figure 1.
Detection of 2-methylthio-cyclic-N6-threonylcarbamoyladenosine (ms2ct6A). (A) The chemical structure of ms2t6A (left) and ms2ct6A (right). The 2-methylthio modification is shown in red, and the N6-threonylcarbamoyl group and its hydantoin form are shown in blue. (B) Secondary structures of B. subtilis tRNALys (left) and T. brucei tRNALys (right) with post-transcriptional modifications: dihydrouridine (D), 5-carboxymethylaminomethyl-2-thiouridine (cmnm5s2U), 2-methylthio cyclic N6-threonylcarbamoyladenosine (ms2ct6A), pseudouridine (Ψ), 7-methylguanosine (m7G), 5-methyluridine (m5U), N2-methylguanosine (m2G), 3-(3-amino-3-carboxypropyl)uridine (acp3U), N2, N2-dimethylguanosine (m2,2G), 2΄-O-methylcytidine (Cm), 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U), 3-(3-amino-3-carboxypropyl)dihydrouridine (acp3D), 5-methylcytidine (m5C). The position numbers of the residues are displayed according to the nucleotide numbering system (44). Pairs of gray triangles in B. subtilis tRNALys indicate the positions of cleavage by RNase T1 that generate RNA fragments containing the anticodon region.