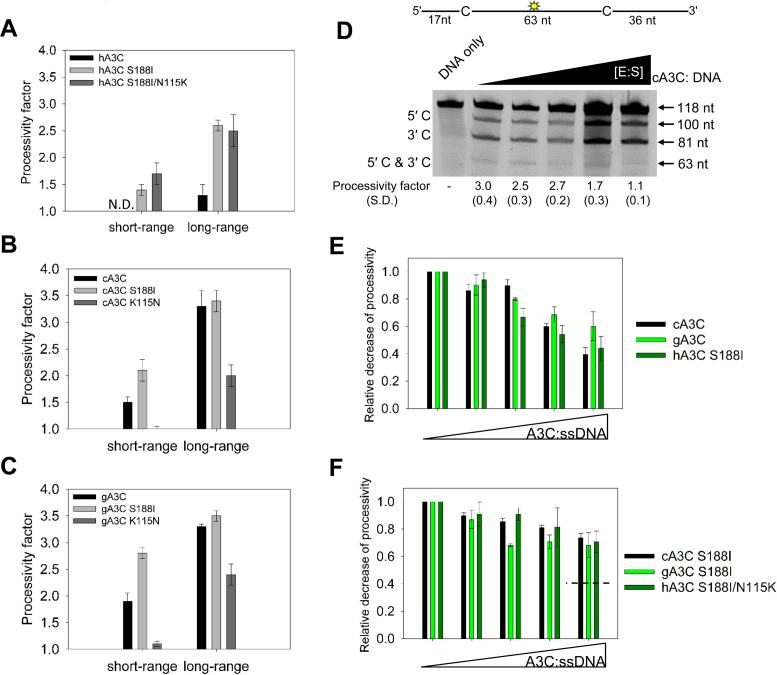
Figure 4.
Dimerization influences processive ssDNA scanning. Processivity of A3C mutants was tested on ssDNA substrates and compared to the wild type enzyme. (A–C) Processivity factor values are shown for short-range movements based on deamination of a 60 nt ssDNA substrate with deamination targets spaced 5 nt apart and long-range movements based on deamination of a 118 nt substrate with deaminated cytosines spaced 63 nt apart for (A) hA3C, hA3C S188I and hA3C S188I/N115K, (B) cA3C, cA3C S188I and cA3C K115N and (C) gA3C, gA3C S188I and gA3C K115N. See Supplementary Figure S4 for a representative gel. (D) Intersegmental transfer ability of cA3C was determined by keeping an A3C/ssDNA ratio of 7:1 constant, but increasing the total reaction components. If the enzyme is able to undergo intersegmental transfer, the assay will result in an apparent decrease in the processivity factor with increasing concentrations of reaction components. The ssDNA substrate contained a fluorescein-labeled deoxythymidine (yellow star) between two deamination targets separated by 63 nt. The measurements of enzyme processivity (processivity factor) and the S.D. are shown below the gel. (E and F) Summary of intersegmental transfer assays shown in Supplementary Figure S5. (E) The monomer/dimer forms of A3C (cA3C, gA3C, hA3C S188I) are better able to undergo intersegmental transfer than the (F) stable dimer forms of A3C (cA3C S188I, gA3C S188I, hA3C S188I/N115K). For comparison, the hatched line in (F) denotes the decrease in processivity observed for monomer/dimer forms of A3C in (E). All values are calculated from at least three independent experiments.