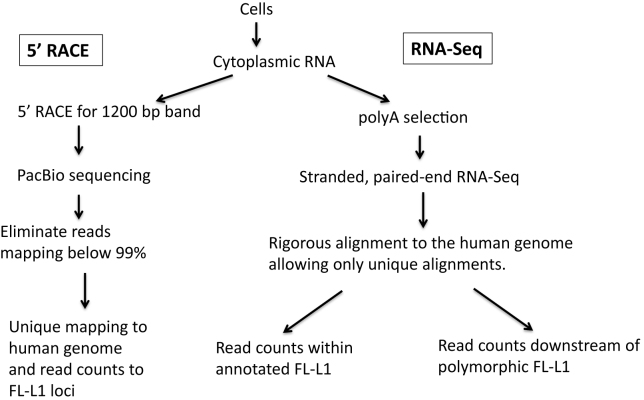
Figure 6.
Flowchart for analysis of expression from full-length L1 loci. In all of our approaches we first isolate cytoplasmic RNAs to eliminate a great deal of the background from L1 elements found in the introns of transcripts in the nucleus. Our 5΄ RACE protocol uses the total cytoplasmic RNA for cDNA synthesis with a L1-specific primer that should generate a 1200 base cDNA only from full-length L1 transcripts that initiate at the beginning of the L1 sequence. This 1200 base cDNA is then amplified using the RACE protocol and subjected to PacBio sequencing. Consensus reads from the PacBio for each molecule are aligned to the human genome and those that align throughout their length with an accuracy lower than 99% are eliminated. This not only eliminates fragments with sequence errors, it also eliminates PCR chimeras between different L1 loci. The accurate L1 alignments are then rigorously aligned to the human genome and only reads that align to one full-length L1 locus better than all others are mapped and counted. In the RNA-Seq studies, the same cytoplasmic RNA is subjected to polyA selection to eliminate rRNA and then subjected to a strand-specific, 2 × 100 bp, paired-end RNA-Seq using the Illumina platform. These paired-end reads are aligned rigorously with BOWTIE, accepting only those alignments where both reads align concordantly at one locus better than anywhere else in the human genome. This alignment is used to either count the reads mapping to each individual full-length L1 locus (left branch) or with reads mapping specifically downstream from known polymorphic L1 loci (right branch) as indirect evidence of expression from those loci.