Abstract
Ribosomes are the protein synthesizing machines of the cell. Recent advances in cryo-EM have led to the determination of structures from a variety of species, including bacterial 70S and eukaryotic 80S ribosomes as well as mitoribosomes from eukaryotic mitochondria, however, to date high resolution structures of plastid 70S ribosomes have been lacking. Here we present a cryo-EM structure of the spinach chloroplast 70S ribosome, with an average resolution of 5.4 Å for the small 30S subunit and 3.6 Å for the large 50S ribosomal subunit. The structure reveals the location of the plastid-specific ribosomal proteins (RPs) PSRP1, PSRP4, PSRP5 and PSRP6 as well as the numerous plastid-specific extensions of the RPs. We discover many features by which the plastid-specific extensions stabilize the ribosome via establishing additional interactions with surrounding ribosomal RNA and RPs. Moreover, we identify a large conglomerate of plastid-specific protein mass adjacent to the tunnel exit site that could facilitate interaction of the chloroplast ribosome with the thylakoid membrane and the protein-targeting machinery. Comparing the Escherichia coli 70S ribosome with that of the spinach chloroplast ribosome provides detailed insight into the co-evolution of RP and rRNA.
INTRODUCTION
Chloroplasts are organelles found in plant and algal cells, which are responsible for carrying out photosynthesis. The origin of chloroplasts is thought to result from an endosymbiotic event where an early eukaryotic cell engulfed a photosynthetic cyanobacterium (1). As such chloroplasts possess their own genome, as well as the transcription and translation machinery to convert the genetic information into polypeptides or proteins (2,3). Chloroplast ribosomes, or chlororibosomes, are very specialized since they are only involved in synthesizing the limited number of proteins encoded in the chloroplast genome (2,3). For example, the complete genome sequence of the Spinacea oleracea (spinach) chloroplast contains 146 genes encoding protein products and structural RNAs (4). The majority of the chloroplast-encoded proteins are targeted to the chloroplast thylakoid membranes and encompass components of the adenosine triphosphate (ATP) synthase, cytochrome b/f and photosystem I and II complexes (4). In addition, chlororibosomes translate NADH dehydrogenase, the large subunit (LSU) of RuBisCO, RNA polymerase subunits and a distinct subset of ribosomal proteins (RPs), 12 from the small subunit (SSU) and 8 from the LSU. Other proteins essential for chloroplast function are nuclear encoded and must therefore be imported into the chloroplast. This includes the remaining 32 chloroplast RPs (cpRPs), which bear N-terminal chloroplast-targeting sequences that are cleaved off upon import (5,6).
Sequence comparisons indicate that the components of the chloroplast translational machinery are similar to those of eubacteria, especially cyanobacteria, but also γ-proteobacteria, such as Escherichia coli. The chloroplast 16S rRNA (cp16S) of the SSU contains 1491 nucleotides (nts) and is therefore only slightly smaller than the E. coli 16S rRNA (Ec16S), which has 1542 nts. The E. coli LSU contains 2 rRNAs, the 5S (120 nts) and 23S (2904 nts) rRNAs, totaling to 3024 nts. While the chloroplast LSU comprises 3 rRNAs, 5S (121 nts), 4.8S (103 nts) and 23S (2810 nts) rRNAs, the total length of 3034 nts is only slightly larger (10 nts) than in E. coli. Similarly, chlororibosomes contain a total of 52 cpRPs (25 in the SSU and 33 in the LSU) and with the exception of L25 and L30, have orthologs in E. coli (5,6). However, the cpRPs are generally larger than their E. coli counterparts, predominantly due to N- and C-terminal extensions (NTEs and CTEs) (5,6). Proteomic studies also identified six non-orthologous proteins, termed ‘plastid-specific RPs’ (or PSRPs) (5–7). Four PSRPs (PSRP1-4) were found to be associated with the SSU and two (PSRP5 and PSRP6) with the LSU (5–7). A cryo-EM reconstruction of the spinach chlororibosome at 9.4 Å provided first insights into the localization of the PSRPs and cpRP extensions (8), however, higher resolution is required to accurately assign and describe the interactions of the PSRPs and cpRP extensions within the chlororibosome.
Here we present a cryo-EM structure of the spinach chlororibosome, with an average resolution of 5.4 Å for the SSU and 3.6 Å for the LSU, revealing the binding site of the PSRP1, PSRP4, PSRP5 and PSRP6 as well as the conformation of numerous cpRP extensions. The structure illustrates how cpRP extensions and PSRPs wind their way through the core of the chlororibosome establishing interactions with neighboring rRNA and RPs. In many cases, the cpRP extensions interact with RNA or protein features that are specific to the chlororibosome, thus providing insight into their co-evolution. We also identify a large conglomerate of cpRP mass adjacent to the tunnel exit site that we suggest facilitates interaction of the chlororibosome with the thylakoid membrane and the protein-targeting machinery.
MATERIALS AND METHODS
Isolation of chloroplast 70S ribosomes
Chloroplast ribosome isolation was performed as described previously (9). Briefly, 6 kg of spinach leaves were deveined and washed thoroughly. The leaves were homogenized (2l/kg of leaves) using 0.7 M Sorbitol in buffer A (10 mM Tris–HCl pH 7.6, 50 mM KCl, 10 mM MgOAc, 7 mM β-mercaptoethanol). The homogenate was filtered through several layers of cheesecloth and one layer of Miracloth (Calbiochem) before centrifugation at 1200 × g for 15 min. The pellet was resuspended in 0.4 M Sorbitol in buffer A and re-centrifuged at 1200 × g for 15 min. The washed chloroplast pellet was resuspended in buffer A supplemented with 2 % (v/v) Triton-X100 and incubated on ice for 30 min. The lysed suspension was clarified by centrifugation at 26 000 × g for 30 min before isolation of crude ribosomes by centrifugation at 50 000 × g for 24 h through a 1M sucrose (in buffer B: buffer A with 10% glycerol). The greenish pellet was washed and then resuspended in buffer B with gentle agitation. The crude ribosomes were clarified by centrifugation at 26 000 × g for 15 min before being either snap frozen at −80°C. Alternatively, the clarified supernatant was applied directly onto a 10–30% sucrose gradient (in buffer B) in order to obtain tight-coupled chloroplast 70S ribosomes.
Negative-stain electron microscopy
Ribosomal particles were diluted in buffer A to a final concentration of 5 A260/ml. One drop of each sample was deposited on a carbon-coated grid. After 30 s, grids were washed with distilled water and then stained with 2% aqueous uranyl acetate for 15 s. The remaining liquid was removed by touching the grid with filter paper. Micrographs were taken using a Morgagni transmission electron microscope (FEI), 80 kV, wide angle 8K CCD at direct magnifications of 110K.
Cryo-electron microscopy and single particle reconstruction
A total of 5 A260/ml chloroplast ribosome sample was applied to 2 nm pre-coated Quantifoil R3/3 holey carbon supported grids and vitrified using a Vitrobot Mark IV (FEI, Eindhoven). Data collection was performed using an FEI Titan Krios transmission electron microscope equipped with a Falcon II direct electron detector (FEI, Eindhoven), using a pixel size of 1.061 Å and an underfocus range of 1.0–2.3 μm resulting in 2031 micrographs. Each micrograph was recorded as a series of 7 frames (3.9 e−/Å2 pre-exposure; 5.2 e−/Å2 dose per frame). All seven frames (accumulated dose of 40.3 e−/Å2) were motion-corrected using the Unblur program (10) and power-spectra, defocus values, astigmatism and estimation of micrograph resolution were determined using CTFFIND4 (11). Five hundred and forty-five micrographs showing Thon rings beyond 3.2 Å resolution were manually inspected further for good areas and power-spectra quality. Three times decimated data were pre-processed using the SPIDER software package (12), in combination with an automated workflow as described previously (13). After initial, automated particle selection based on the program SIGNATURE (14), initial alignment was performed with 56 475 particles using E. coli LSU as a reference structure (15). The dataset could be sorted into 37 626 (66.6%) ribosomal particles and 18 849 (33.3%) non-aligning particles using an incremental K-means-like method of unsupervised 3D sorting (16) (Supplementary Figure S2). Undecimated ribosomal particles were again initially aligned against an E. coli LSU and subsequently refined using FREALIGN (17). Since the SSU of the chlororibosome was flexible, focused alignment and refinement was performed by applying masks either on the SSU or LSU. Due to inherent flexibility, the SSU of the chlororibosome could be refined to an average resolution of 5.4 Å (0.143 FSC) and a local resolution extending to 5.0 Å for the core, whereas the LSU of the chlororibosome could be refined to an average resolution of 3.6 Å (0.143 FSC) and a local resolution extending to <3.5 Å for the core. The local resolution of the final maps was computed using ResMap (18) (Supplementary Figure S2). The final maps were sharpened by dividing the maps by the modulation transfer function of the detector and by applying an automatically determined negative B-factor (−86 for the LSU and −130 for the SSU) to the maps using RELION (19).
Molecular modeling and map-docking procedures
The molecular model of the chloroplast LSU was based on the E. coli-70S-EF-Tu structure (20). The 23S rRNA secondary structure was initially generated by manual alignment of the chloroplast 23S rRNA sequence and the secondary structure map (21) to the E. coli 23S secondary structure map, which shows high structural similarity. The 16S, 5S and 4.8S rRNA sequences of the chloroplast ribosome were aligned accordingly. The resulting rRNA homology models were rigid-body fitted into the respective chloroplast EM-map using Chimera (22). Subsequently, the models were manually adjusted and refined using Coot (23). E. coli-based (20) homology models of the cpRPs were built using SwissModel (24) and HHPred (25) and rigid-body fitted into the map. cpRP-specific extensions were modeled in Coot (23). PSRP5 and PSRP6 were modeled de novo, using secondary structure predictions generated by PsiPred (26) as a reference. The complete atomic model of the chloroplast LSU was subsequently refined using phenix.real_space_refine (27) with secondary structure restraints calculated by PHENIX. In order to reduce the clashscore, the model was additionally refined in reciprocal space using REFMAC (28) in EM mode. Cross-validation against overfitting was performed as described elsewhere (29,30). The statistics of the refined model were obtained using MolProbity (31).
Figure preparation
All figures showing electron densities and atomic models were generated using UCSF Chimera (22) and PyMol Molecular Graphics Systems (version 1.8 Schrödinger).
RESULTS and DISCUSSION
Cryo-EM structure of the chloroplast 70S ribosome
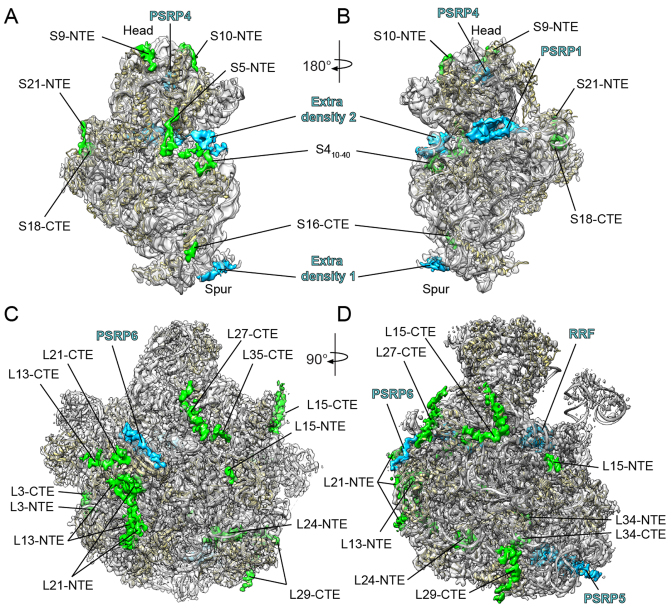
Chloroplast 70S ribosomes were isolated from S. oleracea (spinach) leaves as described previously (8,9) and subjected to single particle cryo-EM analysis. The cryo-EM data was collected on a Titan Krios transmission electron microscope with a Falcon II direct electron detector. From a total of 56,475 ribosomal particles, in silico sorting revealed extreme flexibility of the SSU with respect to the LSU (Supplementary Figure S1). To overcome this conformational heterogeneity, focused alignment was performed independently for each ribosomal subunit using FREALIGN (17). Subsequent refinement yielded cryo-EM reconstructions of the chloroplast SSU and LSU (Figure 1A–D), with an average resolution of 5.4 Å and 3.6 Å, respectively (Supplementary Figure S2 and Table S1).
Figure 1.
Cryo-EM structure of the chloroplast SSU and LSU. (A–D) Cryo-EM map (transparent gray) of the spinach chloroplast (A and B) SSU and (C and D) LSU, illustrating the additional density for cpRPs (green) and extra density assigned to PSRPs (blue) and the ribosome recycling factor (RRF). The molecular model for the SSU and LSU includes rRNA (gray) and cpRPs (yellow).
Analysis of the chloroplast SSU
The resolution of the SSU allowed a homology model of the spinach chloroplast SSU to be rigid body fitted based on the high sequence similarity between the E. coli and S. oleracea rRNA and RPs (8). As already noted (8), the major difference with respect to the 16S rRNA is the shortening of helices h6, h10 and h17 in the chlororibosome rRNA, leading to a truncated spur (Figure 1A and B) when compared to the E. coli SSU. In the previous chlororibosome cryo-EM structure, additional protein density was observed, which was tentatively assigned to PSRP2 and PSRP3, and proposed to compensate for the truncated spur rRNA (8). At higher resolution, this extra spur density was not well-resolved (Figure 1A and B), however, filtering at lower resolution indeed revealed extra density within this region (Supplementary Figure S3). The mass of the extra spur density could not account fully for either PSRP2 or PSRP3, suggesting that if one of these PSRPs is bound there it is highly flexible.
As mentioned, the S. oleracea cpRPs are larger than their respective E. coli counterparts due to the presence of NTEs and/or CTEs (6). To ascertain the location of the cpRP extensions, homology models for the S. oleracea cpRPs were generated based on E. coli templates (20,32), which were then fitted to the cryo-EM map of the chloroplast SSU (Figure 1A and B). In many cases, additional density continuous with the N- or C-termini of the cpRPs could be identified, consistent with the presence of predicted S. oleracea cpRP-extensions that are absent in the respective E. coli RPs (Figure 1A and B). For example, density was observed for the NTE of cpS5, which is 86 aa longer than E. coli S5 (EcS5) (6). In addition, density for the NTEs of cpS9, cpS10 and cpS21 and the CTEs of cpS16 and cpS18 were observed, as well as a rearrangement of the N-terminus of cpS4. The extensions of the cpRPs are located exclusively on the back or cytosolic side of the SSU, but nevertheless encroach on two functional regions related to the path of the mRNA. Specifically, the CTE of cpS18 and the NTE of cpS21 are located at the platform region in vicinity of where the Shine-Dalgarno helix forms between the 5΄ end of the mRNA and the 3΄ end of the 16S rRNA (Supplementary Figure S3). The N-terminus of cpS4, and particularly the NTE of cpS5, surround the mRNA entry channel (Figure 1A and B; Supplementary Figure S3). Curiously, we also observed extra density in this region that does not originate from any of the neighboring cpRP extensions. The extra density connects the head and body of the 30S subunit, namely, bridging the tip of helix h16 in the body with cpS3 of the head. This connection is often referred to as the ‘latch’ because it has been observed to open and close during translation initiation (33,34). Mass spectrometry analysis did not detect additional non-orthologous proteins on the spinach chloroplast SSU (5–7), therefore, the additional density may actually be derived from part of PSRP2 or PSRP3, but we cannot exclude that it is derived from unrelated proteins.
Finally, we identified two additional densities that we assigned to PSRP1 and PSRP4 (Supplementary Figure S4A and B). In agreement with the previous localization (8), we allocated the density within the head of the SSU to PSRP4 (Figure 1A and B) based on its similarity in sequence and binding position with Thx, a small RP identified in the Thermus thermophilus SSU (35). Similarly, we assigned the additional density located within the decoding site on the intersubunit side of the SSU to the N-terminal domain (NTD) of PSRP1, as reported previously (8,36). Sequence alignments indicated that PSRP1 is not a bone fide cpRP but rather a homolog of a long form hibernation-promoting factor, which is responsible for 100S formation (70S dimerization) (37). The NTD of PSRP1 is homologous with YfiA and the short form HPF, both of which have also been shown to bind analogously to the SSU of bacterial 70S ribosomes (38,39), overlapping the binding site of the mRNA and tRNAs in the A- and P-sites (Supplementary Figure S4C and D). No density was observed for the C-terminal domain of PSRP1, which has been shown to be responsible for 100S formation in some bacteria (40,41).
Molecular model for the chloroplast LSU
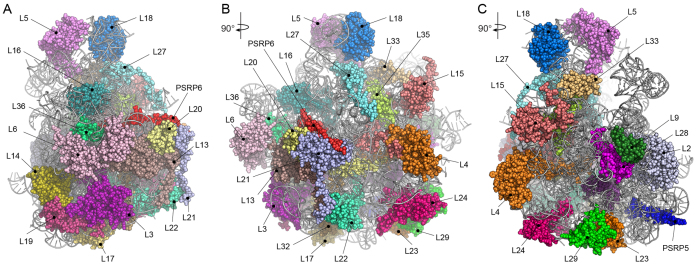
Consistent with the local resolution calculations (Supplementary Figure S2), the electron density was particularly well resolved within the core of the LSU, whereas the periphery of the subunit was less defined. We were able to generate molecular models for 28 of the 33 cpRPs present in the chlororibosome (Figure 2A–C; Supplementary Table S2). cpRPs L1, L10, L11, L7/L12 and L31 were not modeled due to poor density. The density for cpL5, cpL6 and cpL18 allowed only a rigid body fit of a homology model based on EcL5 and EcL6, and only the NTD of cpL9 was included in the final model. As observed previously (8), density was not observed for L25 and L30, consistent with the absence of genes encoding these cpRPs in plant and chloroplast genomes (4). We could also model domain I of the chloroplast ribosome recycling factor (cpRRF) (Supplementary Figure S4), which was bound analogously to that reported previously on the chlororibosome at lower resolution (8) as well as on bacterial ribosomes (42,43). Together with cpEF-G, cpRRF has been demonstrated to dissociate PSRP1 from the chlororibosome (36). In addition, molecular models are presented for the complete 5S and 2843 (97.6%) of the 2913 nucleotides that comprise the 4.8S and 23S rRNAs (Figure 3A and B; Supplementary Figure S5).
Figure 2.
Molecular model for the chloroplast LSU. (A–C) Three overviews of the chloroplast LSU with rRNA (gray ribbons) and modeled cpRPs and PSRPs shown with a spacefill representation and colored and labeled individually.
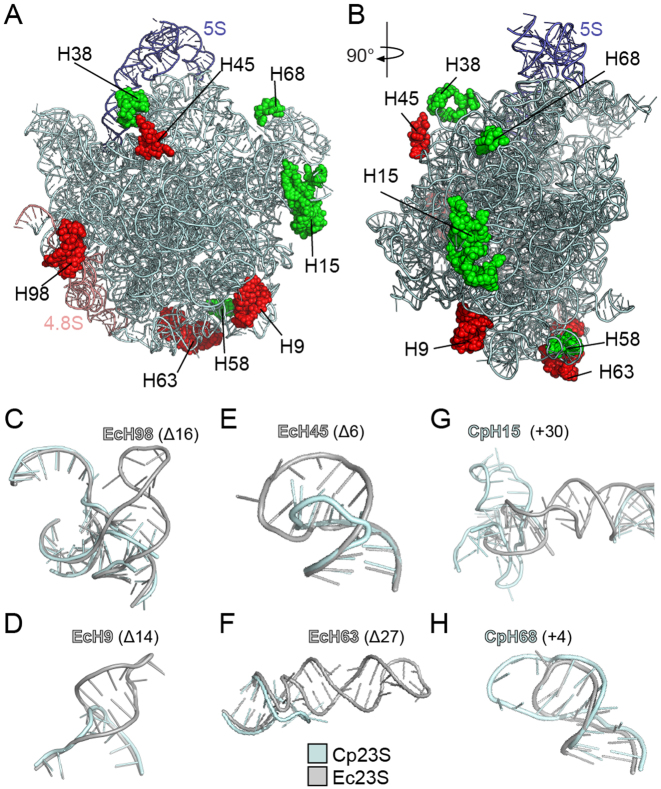
Figure 3.
Location of rRNA insertions and deletions in the chlororibosome. (A and B) Two overviews of the chloroplast LSU with 4.8S (teal), 5S (purple) and 23S (cyan) rRNA, highlighting insertions (green) on the cp23S and deletions (red) relative to Ec23S. (C–F) Examples of deletions in the cp23S (cyan) relative to the Ec23S (gray) include deletion of (C) 16 nts in H98, (D) 14 nts in H9, (E) 6 nts in H45 and (F) 27 nts in H63. (G and H) Examples of additions in the cp23S (cyan) relative to the Ec23S (gray) include insertion of (G) 30 nts in H15 and (H) 4 nts in H68.
Features of the chloroplast LSU rRNAs
Unlike the mammalian mitoribosome where a tRNA molecule substitutes for the lack of a 5S rRNA (44,45), the chlororibosome contains a 5S rRNA (Figure 3A and B) that is highly similar in sequence and structure to the bacterial 5S rRNA. As mentioned, the chloroplast 23S rRNA is present in the chlororibosome as two pieces, a 5΄ fragment representing H1-H97 of domains I-VI (hereafter referred to as cp23S rRNA) and a 3΄ fragment comprising H99-H101 of domain VII (termed 4.8S rRNA) (Figure 3A and Supplementary Figure S5). This results in the loss of H98 (Δ16 nts) that links domains VI and VII within the E. coli 23S (Ec23S) rRNA (Figure 3C). Together with reductions in helices H9 (Δ14 nts), H45, (Δ6 nts), H63 (Δ27 nts) (Figure 3A and B), the cp23S rRNA has a total of 75 nts missing relative to the Ec23S rRNA. While the reductions lead to a shortening in the length of H9 and H45 (Figure 3D and E), the effect on H98 and H63 results in the complete absence of these helices in the chlororibosome (Figure 3C and F). Nevertheless, the combined length of the chloroplast LSU rRNA (3034 nts) is similar to that for E. coli (3024 nts) because the four rRNA reductions in the cp23S rRNA are compensated by five rRNA additions (8). This includes additional nucleotides within H15 (+30 nts), H38 (+20 nts), H58 (+23 nts) and H68 (+4 nts) of the cp23S rRNA, as well as +8 nt insertion in the linker connecting H100 and H101 of the 4.8S rRNA (Figure 3A and B). The insertions within H15 (Figure 3G) and H68 (Figure 3H) are base-paired and well resolved in the model, whereas the density for the non-base-paired insertions within H38 and the H100–H101 linker are poorly defined and therefore not included in the final model.
Localization of cpRPs and extensions
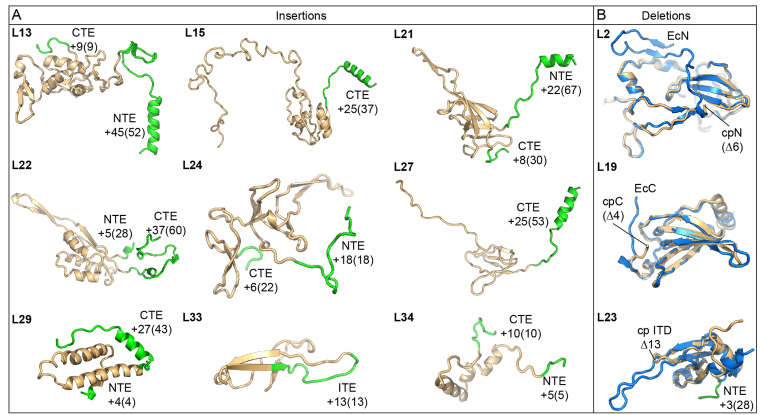
In contrast to the SSU cpRPs, the LSU cpRPs are significantly longer than their E. coli counterparts due to the presence of NTEs and/or CTEs (5). As expected, additional density continuous with the N- or C-termini of the LSU cpRPs was observed (Figure 1C and D), allowing 283 amino acids of the cpRP extensions to be modeled (Figure 4A; Supplementary Table S2). In particular, cpRPs L13, L15, L21, L22, L24, L27, L29 and L34 have long NTEs and/or CTEs (Figure 4A). In addition, cpL33 has a β-hairpin with an internal expansion of 13 aa compared to EcL33 (Figure 4A). There are four major exceptions of cpRPs (L2, L17, L19 and L23) that have significant deletions (>2 aa) compared to their E. coli counterparts. In the chlororibosome, cpL17 is C-terminally truncated by 11 aa, although only 4 aa of these are observed in the E. coli 70S ribosome structures (20,32). The N-terminus of cpL2 is shorter than EcL2 by only 6 aa, yet we observed no density for the first 25 aa (Figure 4B). Similarly, the CTE of cpL19 is not only 4 aa shorter than EcL19, but the last 11 aa also adopt a distinct conformation (Figure 4B). Lastly, the β-hairpin of EcL23 that reaches into the tunnel lumen in the E. coli ribosome (20,32) is significantly shorter in the chlororibosome (Figure 4B), reminiscent of the archaeal/eukaryotic L23 homologs (46,47). However, unlike archaeal/eukaryotic ribosomes that compensate for the truncated L23 with the presence of aeL38 (46,47), the equivalent space remains vacant in the chlororibosome.
Figure 4.
Molecular models indicating cpRP extensions and deletions. (A) Structures of cpRPs showing the core region equivalent to the respective EcRPs (gold) with N-terminal extensions (NTEs), C-terminal extensions (CTEs) or the internal expansion (ITE) highlighted (green). The numbers indicate the modeled residues with the total expansion length indicated in parentheses. (B) Structures of cpRPs (gold) compared with the respective EcRPs (blue) highlighting amino acid deletions (in parentheses) in cpRPs relative to EcRPs.
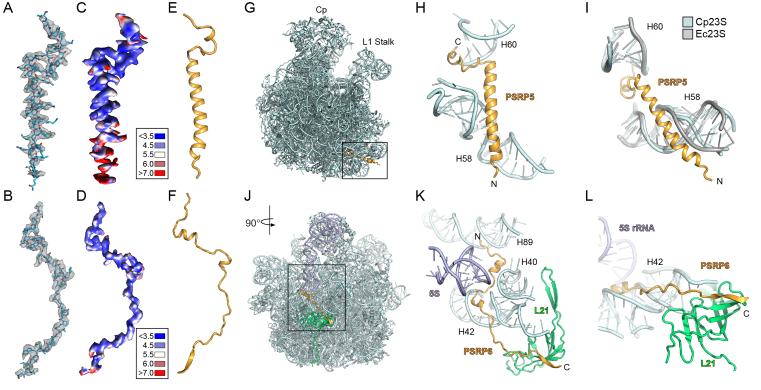
Binding sites of PSRP5 and PSRP6 on the LSU
Subsequent to modeling of the LSU rRNAs as well as all the cpRPs and cpRP extensions, we noticed that two unmodeled regions of electron density were present in the cryo-EM map, which we assigned to PSRP5 and PSRP6 (Figure 1C and D). Due to their buried location within the chlororibosome, the electron density was well resolved (Figure 5A and B), in agreement with local resolution calculations (Figure 5C and D), enabling unambiguous models for both PSRP5 and PSRP6 to be generated (Figure 5E and F; Supplementary Figure S4). Consistent with secondary structure predictions, PSRP5 consists of a short C-terminal α-helix connected by a linker to a long central α-helix (Figure 5E). The binding site of PSRP5 is located at the base of the LSU directly under the L1 stalk, with the N-terminus extending toward the intersubunit interface (Figure 5G). We note that 38 aa are missing from the N-terminus in our model, presumably due to flexibility outside of the ribosome. The surface of the buried regions of PSRP5 is highly positively charged (Supplementary Figure S4F and G), as would be expected from the surrounding negatively charged rRNA environment. The short C-terminal α-helix of PSRP5 inserts into the minor groove of H60, whereas the central α-helix establishes interactions with H58 (Figure 5H). The specificity of PSRP5 for the chlororibosome may be due to the interaction with H58, since there are significant differences in both the sequence and structural conformation of H58 when comparing with the E. coli 70S ribosome (Figure 5I). We note that the position of PSRP5 was mis-assigned in the previous structure of S. oleracea chloroplast 70S ribosome (8), probably due to the small size of the protein and the limited resolution of the reconstruction.
Figure 5.
Localization of PSRP5 and PSRP6 on the chlororibosome. (A and B) Cryo-EM electron density (mesh) with molecular models for (A) PSRP5 and (B) PSRP6. (C and D) Cryo-EM electron density for (C) PSRP5 and (D) PSRP6 colored according to local resolution. (E and F) Molecular models showing secondary structure for (E) PSRP5 and (F) PSRP6. (G) Binding site of PSRP5 (gold) on the LSU (cyan). (H and I) Interaction between PSRP5 (gold) and H58 and H60 of the cp23S rRNA (cyan), with (I) comparison of different conformation of H58 from the Ec23S rRNA (gray). (J) Binding site of PSRP6 (gold) on the LSU (cyan). (K and L) Interaction between the N-terminus of PSRP6 (gold) and H40, H42 and H89 of the cp23S rRNA (cyan), and (L) the C-terminus of PSRP6 (gold) with the cpL21 (green).
PSRP6 adopts a very extended conformation (Figure 5F) that winds its way through the ribosome (Figure 5J–L). The N-terminal half of PSRP6 is predominantly positively charged (Supplementary Figure S4I–K), consistent with the extensive interaction with the negatively charged rRNA (Figure 5K). The N-terminus of PSRP6 interacts with the minor grooves of H89, H40 and H42 as it winds its way out of the ribosomal core (Figure 5K). The two short central α-helices of PSRP6 are positioned within the minor grooves of H40 and H42 and are separated by a linker region that passes near to the 5S rRNA (Figure 5K). The C-terminal half of PSRP6 is less charged (Supplementary Figure S4I), consistent with an interaction with the globular domain of cpL21, rather than with rRNA. The C-terminus of PSRP6 donates a β-strand to augment the β-sheet of cpL21 (Figure 5L) before extending into the solvent where the C-terminal 22 aa are not visualised. The conservation of this region between S. oleracea chloroplast and E. coli 70S ribosomes suggests that PSRP6 could in principal bind analogously to the E. coli 70S ribosome.
cpRP extensions and rRNA stabilization
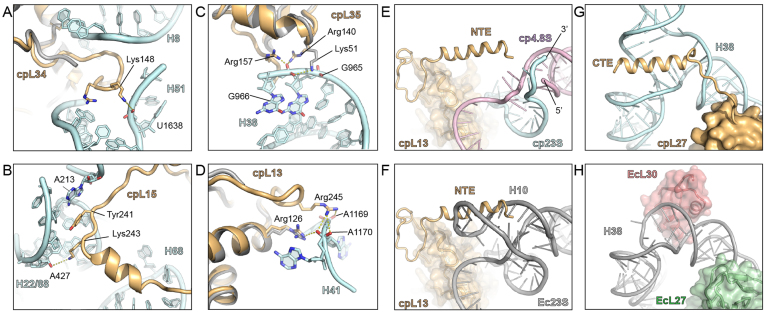
Generally, the NTE and CTE of cpRPs contain positively charged amino acids that establish additional interactions with the surrounding rRNA, predominantly with the phosphate-oxygens of the backbone. For example, the 10 aa CTE of cpL34 interacts with the loop of helix H8 of the 23S rRNA and forms a potential hydrogen bond from Lys148 with the backbone of U1638 within H51 (Figure 6A). In many cases, the cpRP extensions interact with the minor groove of rRNA helices. Such an interaction is illustrated by the 25 aa CTE of cpL15, which inserts into the minor groove of a helix formed from the loops of H22 and H88 (Figure 6B). Lys243 comes within hydrogen bonding distance of the ribose of A427 and Tyr241 stacks upon A213 that makes an A-minor interaction within the H22/H88 helix (Figure 6B). Similarly, the 18 aa NTE of cpL24 that penetrates deeper into the ribosomal core, approaches the minor groove of an rRNA helix formed from the loops of H6 and H7, before the N-terminus emerges within the tunnel lumen (see later).
Figure 6.
Interaction of cpRP extensions with rRNA. (A–D) Examples of interaction of cpRP extensions (gold) with cp23S rRNA (cyan) include the (A) CTE of cpL34 with H51, (B) CTE of cpL15 with H22/H88, (C) CTE of cpL35 with H38 and (D) NTE of cpL13 with H41. (E) Interaction of NTE of cpL13 (gold) with the 3΄ end of the cp23S (cyan) and the 5΄ end of the 4.8S (pink) in the chlororibosome, superimposed with the (F) Escherichia coli 70S ribosome showing that H10 of the Ec23S (gray) overlaps with the NTE of cpL13 (gold). (G) Interaction of CTE of cpL27 (gold) with the three-way junction of H38 (cyan) of the chlororibosome, whereas in the (H) E. coli 70S ribosome, EcL27 (green) has no extension and H38 (gray) is bound by EcL30 (red).
We also observed that the cpRP extensions often reinforce interactions with rRNA elements that are already contacted by the core of the cpRP, as illustrated by cpRPs L35 and L13 (Figure 6C and D). Arg140 in the core of cpL35 contacts the phosphate-oxygen of G966 in H38, an interaction also observed for EcL35 (Figure 6C). This contact is reinforced in the chlororibosome by a potential hydrogen bond from Arg157 within the 7 aa CTE of cpL35 to the backbone of C966 within H38 (Figure 6C). Similarly, the interaction from Arg126 in the core of cpL13 with A1170 in H41 is reinforced in the chlororibosome by an additional hydrogen bond from Arg245 within the NTE of cpL13 to the backbone of A1170 within H41 (Figure 6D).
Three of the cpRP extensions contain α-helical secondary structure, namely within the NTE of cpL13 and the CTEs of cpL15 and cpL27 (Figure 4A). The α-helix within the CTE of cpL15 interacts with H68, which as mentioned is extended in the chlororibosome compared to the E. coli 70S (Figure 3H). In the chlororibosome, the NTE of cpL13 forms an α-helix that interacts with the junction where the 5΄ end of the 4.8S rRNA meets the 3΄ end of the cp23S rRNA (Figure 6E). Comparison with the E. coli 70S ribosome revealed that the N-terminal α-helix of cpL13 occupies the position of H10 of the Ec23S rRNA (Figure 6F), which is absent in the chlororibosome (Figure 6E). The α-helix within the CTE of cpL27 appears to stabilize a three-way junction formed by the insertion of 20 nts within H38 of the cp23S rRNA (Figure 6G), which is lacking in the Ec23S rRNA (Figure 6H). The site of insertion in H38 in the cp23S rRNA correlates with the position of expansion segment 12 (ES12L) in eukaryotic 80S ribosomes (47,48). In the E. coli ribosome, EcL30 contacts H38 in the vicinity of the insertion site (Figure 6H). Such an L30-H38 interaction would not be possible in the chlororibosome due to the presence of the additional rRNA helix in H38, thus providing a possible explanation as to why L30 is missing in plant chloroplasts.
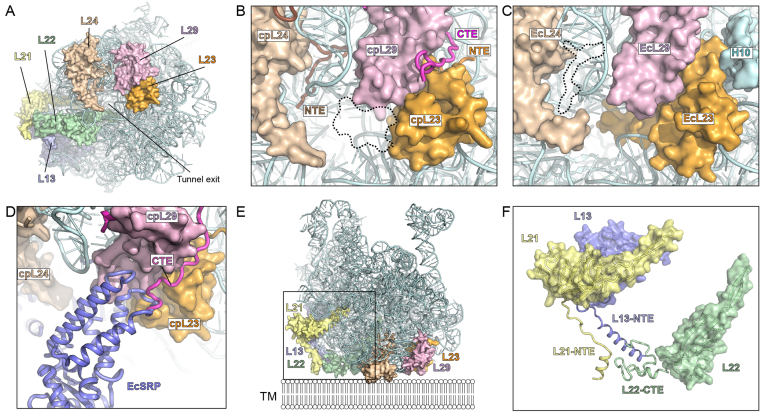
Intertwining of cpRP extensions at the tunnel exit
A number of differences with the E. coli 70S ribosome were evident when examining the back or cytosolic side of the LSU of the chlororibosome, in particular, the region surrounding the tunnel exit site. As mentioned, the β-hairpin of cpL23 is shorter than EcL23 leading to an enlarged luminal space near the exit site of the chlororibosome (Figure 7A–C). In contrast, the opposite side of the tunnel from cpL23 has extra mass due to the presence of the NTE of cpL24 that penetrates into the ribosomal core from the surface located globular domain (Figure 7B). The 27 aa CTE of cpL29 intertwines with the NTE of cpL23 (Figure 7B), which together occupy the space where 23S rRNA helix H10 is situated in the E. coli 70S ribosome (Figure 7C). Comparison with the binding site of E. coli SRP on the ribosome (49,50), suggests that the CTE of cpL29 could play a role in recruitment cpSRP54 to the chlororibosome (Figure 7D).
Figure 7.
Interaction of cpRP extensions with rRNA. (A) View onto the tunnel exit site of the chloroplast LSU with rRNA (cyan) and highlighting cpRPs L23 (orange), L29 (purple), L24 (tan), L22 (green), L13 (blue) and L21 (yellow). (B) Zoom of (A) highlighting the NTE of cpL24 and cpL23, and the CTE of cpL29 as well as the shorter β-hairpin of cpL23. (C) Equivalent view of (B) but for Escherichia coli 70S ribosome, highlighting the absence of EcL24-NTE and the presence of the β-hairpin of EcL23 in the tunnel lumen, as well as H10 of Ec23S rRNA. (D) Superimposition of EcSRP (blue) on chlororibosome illustrating overlap with the CTE of cpL29. (E) Chloroplast LSU, colored as in (A), illustrating additional cpRP protein mass that expands the potential surface area of the LSU and facilitates its possible interaction with the thylakoid membrane (TM). (F) Zoom of boxed region in (E) without rRNA to illustrate the contribution of the cpRP extensions (NTE/CTE) of L21 (yellow), L13 (blue) and L22 (green) to the thylakoid membrane interaction surface.
By far the largest conglomerate of cpRP extensions is located at the back of the LSU adjacent to the tunnel exit site (Figure 7E). This conglomerate comprises the 45 aa (of 52 aa) NTE of cpL13, 22 aa (of 67 aa) from the NTE of cpL21 and 37 aa (of 60 aa) CTE of cpL22, which reach out from the respective globular domains to form multiple protein–protein interactions with each other (Figure 7F). The high flexibility of the extensions, and the poor quality of the density at the periphery of the ribosome, enabled only the backbone of the protein extensions to be traced. Moreover, the N-terminal 45 aa of the NTE of cpL21 could not be modeled, although density was observed at lower thresholds suggesting that these residues establish additional interactions with the CTE of cpL22. Collectively, these cpRP extensions expand the area of the LSU and could facilitate interaction with the thylakoid membrane (Figure 7E).
CONCLUSION
Here we present a near-complete molecular model for the spinach chloroplast LSU, revealing the location of rRNA insertions and deletions, cpRP extensions as well as the binding site of two plastid-specific RPs, PSRP5 and PSRP6. Prior to submission, a cryo-EM structure of the spinach chloroplast 50S subunit was reported by Ahmed and coworkers (51). Generally, the results appear to be in good agreement with our structure, although a careful comparison cannot be undertaken as the cryo-EM map and model were not yet available at the time of submission, nor during the review process. In general, the differences of the chlororibosome with respect to the eubacterial E. coli 70S ribosome are localized to peripheral regions of the ribosome and not within core functional regions that would be expected to influence translational activity, such as the subunit interface, peptidyl-transferase center or translation factor binding site. One major exception is related to the ribosomal tunnel through which the nascent polypeptide chain passes as it is synthesized. In the chlororibosome, we observed that the lower region of the tunnel differs from bacteria due to a shorter β-hairpin of cpL23 and the additional presence of the NTE of cpL24. Formation of α-helical secondary structure within nascent polypeptides chains has been observed in this region of the ribosomal tunnel (52). Structural changes within this region of the chlororibosome may facilitate targeting and insertion of transmembrane-containing proteins into the thylakoid membrane. In this respect, we also note that the CTE of cpL29 could play a role in recruitment of cpSRP54 to the chlororibosome. Unlike bacterial SRPs, the cpSRP lacks the 4.5S RNA (termed, SRP RNA) and comprises only the SRP54 protein, and therefore the CTE of cpL29 may contribute to stabilization of SRP54 interaction with the chlororibosome. Finally, we observed a large conglomerate of cpRP extensions that expand the surface area at the back of the LSU. We suggest that this may facilitate interaction of the chlororibosome directly with the thylakoid membrane and/or membrane-bound components of the targeting machinery, and thereby increase the efficiency of membrane protein insertion. As mentioned, the majority of the chloroplast-encoded proteins is targeted to the thylakoid membranes, including components of the ATP synthase, cytochrome b/f and especially photosystem I and II complexes (4).
ACCESSION NUMBERS
The coordinates and cryo-EM map for the chlororibosome have been deposited in the Protein Data Bank and EM DataBank under accession codes PDB ID 5MLC and EMD-3525/EMD-3526, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Heidemarie Sieber, Susi Rieder and Charlotte Ungewickell for expert technical assistance and Sarah Schlaak for help with data analysis. CIISB research infrastructure project LM2015043 funded by MEYS CR is gratefully acknowledged for the financial support of the measurements at the CF Cryo-electron Microscopy and Tomography CEITEC MU.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [FOR-1805, SPP-1879, GRK-1721 to D.N.W.]; iNEXT project [653706]; Horizon 2020 program of the European Union; MEYS CR (CIISB research infrastructure project LM2015043. Funding for open access charge: DFG.
Conflict of interest statement. None declared.
REFERENCES
- 1.Reyes-Prieto A., Weber A.P., Bhattacharya D.. The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 2007; 41:147–168. [DOI] [PubMed] [Google Scholar]
- 2.Tiller N., Bock R.. The translational apparatus of plastids and its role in plant development. Mol. Plant. 2014; 7:1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun Y., Zerges W.. Translational regulation in chloroplasts for development and homeostasis. Biochim. Biophys. Acta. 2015; 1847:809–820. [DOI] [PubMed] [Google Scholar]
- 4.Schmitz-Linneweber C., Maier R.M., Alcaraz J.P., Cottet A., Herrmann R.G., Mache R.. The plastid chromosome of spinach (Spinacia oleracea): complete nucleotide sequence and gene organization. Plant Mol. Biol. 2001; 45:307–315. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi K., Subramanian A.R.. The plastid ribosomal proteins. Identification of all the proteins in the 50S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 2000; 275:28466–28482. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi K., von Knoblauch K., Subramanian A.R.. The plastid ribosomal proteins. Identification of all the proteins in the 30 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 2000; 275:28455–28465. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi K., Subramanian A.R.. Proteomic identification of all plastid-specific ribosomal proteins in higher plant chloroplast 30S ribosomal subunit. Eur. J. Biochem. 2003; 270:190–205. [DOI] [PubMed] [Google Scholar]
- 8.Sharma M.R., Wilson D.N., Datta P.P., Barat C., Schluenzen F., Fucini P., Agrawal R.K.. Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:19315–19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartsch M., Kimura M., Subramanian A.R.. Purification, primary structure, and homology relationships of a chloroplast ribosomal protein. Proc. Natl. Acad. Sci. U.S.A. 1982; 79:6871–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant T., Grigorieff N.. Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. Elife. 2015; 4:e06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohou A., Grigorieff N.. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015; 192:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A.. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996; 116:190–199. [DOI] [PubMed] [Google Scholar]
- 13.Becker T., Franckenberg S., Wickles S., Shoemaker C.J., Anger A.M., Armache J.P., Sieber H., Ungewickell C., Berninghausen O., Daberkow I. et al. . Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012; 482:501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J.Z., Grigorieff N.. SIGNATURE: a single-particle selection system for molecular electron microscopy. J. Struct. Biol. 2007; 157:168–173. [DOI] [PubMed] [Google Scholar]
- 15.Arenz S., Meydan S., Starosta A.L., Berninghausen O., Beckmann R., Vazquez-Laslop N., Wilson D.N.. Drug sensing by the ribosome induces translational arrest via active site perturbation. Mol. Cell. 2014; 56:446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loerke J., Giesebrecht J., Spahn C.M.. Multiparticle cryo-EM of ribosomes. Methods Enzymol. 2010; 483:161–177. [DOI] [PubMed] [Google Scholar]
- 17.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J. Struct. Biol. 2007; 157:117–125. [DOI] [PubMed] [Google Scholar]
- 18.Kucukelbir A., Sigworth F.J., Tagare H.D.. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014; 11:63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer N., Neumann P., Konevega A.L., Bock L.V., Ficner R., Rodnina M.V., Stark H.. Structure of the E. coli ribosome-EF-Tu complex at <3 A resolution by C-corrected cryo-EM. Nature. 2015; 520:567–570. [DOI] [PubMed] [Google Scholar]
- 21.Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Muller K.M. et al. . The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. Biomed Central Bioinformatics. 2002; 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 24.Guex N., Peitsch M.C.. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997; 18:2714–2723. [DOI] [PubMed] [Google Scholar]
- 25.Soding J., Biegert A., Lupas A.N.. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005; 33:W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchan D., Minneci F., Nugent T., Bryson K., Jones D.. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013; 41:W340–W348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vagin A.A., Steiner R.A., Lebedev A.A., Potterton L., McNicholas S., Long F., Murshudov G.N.. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2184–2195. [DOI] [PubMed] [Google Scholar]
- 29.Brown A., Long F., Nicholls R.A., Toots J., Emsley P., Murshudov G.. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 2015; 71:136–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amunts A., Brown A., Bai X.C., Llacer J.L., Hussain T., Emsley P., Long F., Murshudov G., Scheres S.H., Ramakrishnan V.. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014; 343:1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noeske J., Wasserman M.R., Terry D.S., Altman R.B., Blanchard S.C., Cate J.H.. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 2015; 22:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Llacer J.L., Hussain T., Marler L., Aitken C.E., Thakur A., Lorsch J.R., Hinnebusch A.G., Ramakrishnan V.. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol. Cell. 2015; 59:399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hussain T., Llacer J.L., Wimberly B.T., Kieft J.S., Ramakrishnan V.. Large-scale movements of IF3 and tRNA during bacterial translation initiation. Cell. 2016; 167:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wimberly B.T., Brodersen D.E., Clemons W.M., Morgan-Warren R.J., Carter A.P., Vonrhein C., Hartsch T., Ramakrishnan V.. Structure of the 30S ribosomal subunit. Nature. 2000; 407:327–339. [DOI] [PubMed] [Google Scholar]
- 36.Sharma M.R., Donhofer A., Barat C., Marquez V., Datta P.P., Fucini P., Wilson D.N., Agrawal R.K.. PSRP1 is not a ribosomal protein, but a ribosome-binding factor that is recycled by the ribosome-recycling factor (RRF) and elongation factor G (EF-G). J. Biol. Chem. 2010; 285:4006–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida H., Wada A.. The 100S ribosome: ribosomal hibernation induced by stress. Wiley interdisciplin. Rev. RNA. 2014; 5:723–732. [DOI] [PubMed] [Google Scholar]
- 38.Vila-Sanjurjo A., Schuwirth B.S., Hau C.W., Cate J.H.D.. Structural basis for the control of translational initiation during stress. Nat. Struct. Mol. Biol. 2004; 11:1054–1059. [DOI] [PubMed] [Google Scholar]
- 39.Polikanov Y.S., Blaha G.M., Steitz T.A.. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012; 336:915–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puri P., Eckhardt T.H., Franken L.E., Fusetti F., Stuart M.C., Boekema E.J., Kuipers O.P., Kok J., Poolman B.. Lactococcus lactis YfiA is necessary and sufficient for ribosome dimerization. Mol. Microbiol. 2014; 91:394–407. [DOI] [PubMed] [Google Scholar]
- 41.Basu A., Yap M.N.. Ribosome hibernation factor promotes Staphylococcal survival and differentially represses translation. Nucleic Acids Res. 2016; 44:4881–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson D.N., Schluenzen F., Harms J.M., Yoshida T., Ohkubo T., Albrecht R., Buerger J., Kobayashi Y., Fucini P.. X-ray crystallography study on ribosome recycling: the mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J. 2005; 24:251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weixlbaumer A., Petry S., Dunham C.M., Selmer M., Kelley A.C., Ramakrishnan V.. Crystal structure of the ribosome recycling factor bound to the ribosome. Nat. Struct. Mol. Biol. 2007; 14:733–737. [DOI] [PubMed] [Google Scholar]
- 44.Amunts A., Brown A., Toots J., Scheres S.H., Ramakrishnan V.. Ribosome. The structure of the human mitochondrial ribosome. Science. 2015; 348:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greber B.J., Bieri P., Leibundgut M., Leitner A., Aebersold R., Boehringer D., Ban N.. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015; 348:303–308. [DOI] [PubMed] [Google Scholar]
- 46.Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A.. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000; 289:905–920. [DOI] [PubMed] [Google Scholar]
- 47.Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M.. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011; 334:1524–1529. [DOI] [PubMed] [Google Scholar]
- 48.Anger A.M., Armache J.P., Berninghausen O., Habeck M., Subklewe M., Wilson D.N., Beckmann R.. Structures of the human and Drosophila 80S ribosome. Nature. 2013; 497:80–85. [DOI] [PubMed] [Google Scholar]
- 49.Halic M., Blau M., Becker T., Mielke T., Pool M.R., Wild K., Sinning I., Beckmann R.. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006; 444:507–511. [DOI] [PubMed] [Google Scholar]
- 50.Jomaa A., Boehringer D., Leibundgut M., Ban N.. Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat. Commun. 2016; 7:10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahmed T., Yin Z., Bhushan S.. Cryo-EM structure of the large subunit of the spinach chloroplast ribosome. Sci. Rep. 2016; 6:35793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson D.N., Beckmann R.. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr. Opin. Struct. Biol. 2011; 21:1–10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.