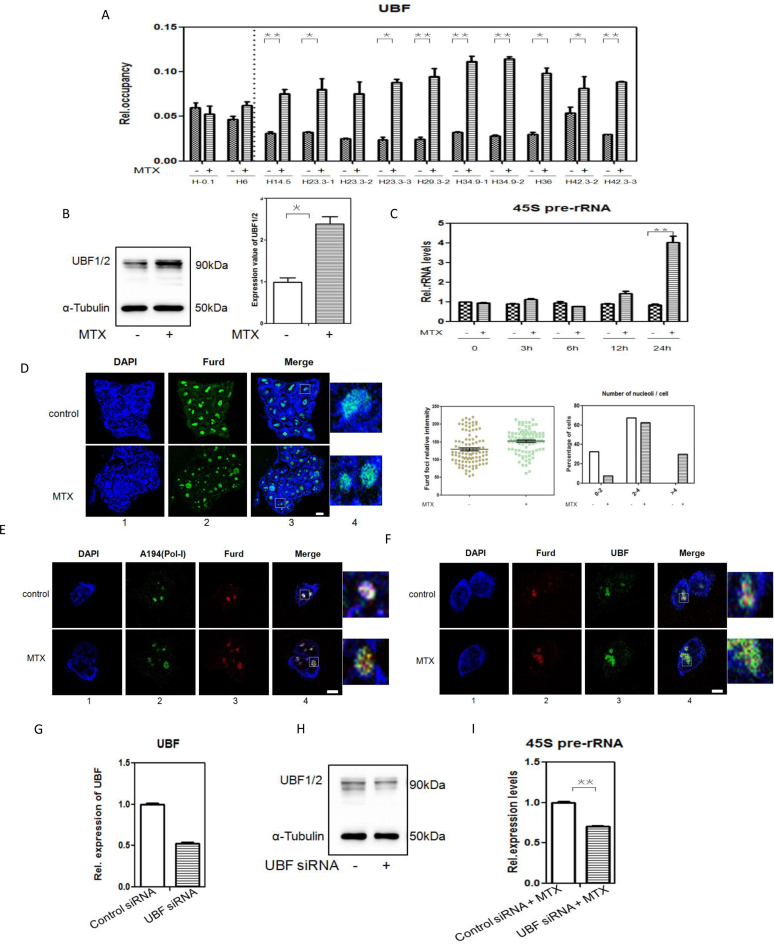
Figure 5.
MTX promotes rRNA transcription and UBF specifically binds to DSB regions of rRNA genes (A) Binding of UBF to the DSBs in rRNA genes unit after 0.12 μM MTX treatment compared with the control, determined by ChIP-qPCR analysis. *P <0.05; **P <0.01. (B) Detection of UBF expression in mESCs cultured in complete medium and complete medium with 0.12 μM MTX treatment, by western blotting. α-Tubulin was used as a loading control. Positions of molecular weight markers are indicated. *P <0.05. (C) Expression levels of 45S pre-rRNA in mESCs cultured in complete medium with 0.12 μM MTX for 3, 6, 12 or 24 h, determined by RT-qPCR. The assays were performed in triplicate and the mean ± SD was calculated. **P <0.01. (D) Detection of ongoing rRNA synthesis by FUrd incorporation assays on mESCs subjected to 0.12 μM MTX for 24 h. Transcription was monitored by a FUrd pulse to observe incorporation into nascent nucleolar transcripts. FUrd detection with BrdU antibody was by confocal microscopy (panel 2). Nuclear DNA was stained by DAPI (panel 1). Merged images (panel 3) are shown. One representative nucleus with associated merged images (panel 4) are shown. Green: FUrd; blue: DAPI staining. Bar, 5 μM. Relative intensity of FUrd foci and the number of nucleoli per cell with FUrd foci were analyzed. The histogram shows the average number of nucleoli per cell based on an analysis of 93 cells from two independent experiments. (E) Co-localization between nucleolar proteins and rRNA transcripts by double-stained preparations of nucleolar markers (panel 2, Pol-I largest subunit A194) and FUrd pulse (panel 3, BrdU antibody).Nuclear DNA was stained by DAPI(panel 1).Merged images (panel 4) are shown. Green: A194(Pol-I); red: FUrd; blue: DAPI staining. Bar, 5 μM. (F) Co-localization between rRNA transcripts and UBF by double-stained preparations of FUrd pulse (panel 2, BrdU antibody) and UBF(panel3, upstream binding factor antibody).Nuclear DNA was stained by DAPI (panel 1). Merged images (panel 4) are shown. Green: UBF; red: FUrd; blue: DAPI staining. Bar, 5 μM. (G) Efficiency of siRNA mediated UBF depletion on mRNA level. The levels of the mRNAs UBF were analysed in mESCs with either a non-targeting siRNA (control siRNA) or siRNA directed against UBF(UBF siRNA). The assays were performed in triplicate and the mean ± SD was calculated. (H) Efficiency of siRNA mediated UBF depletion on protein level. mESCs were transfected with either control siRNA or UBF siRNA and UBF1/2 were determined 72 h post-transfection by Western-blotting. α-Tubulin was used as a loading control. Positions of molecular weight markers are indicated. (I) Expression levels of 45S pre-rRNA in mESCs with control siRNA or UBF siRNA cultured in complete medium with 0.12 μM MTX for 24 h, determined by RT-qPCR. The assays were performed in triplicate and the mean ± SD was calculated. **P <0.01.