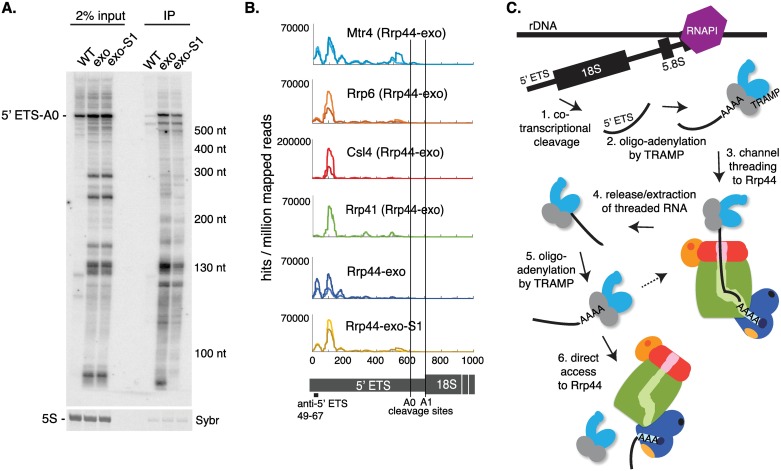
Fig 2. Targeting of the pre-rRNA 5’ external transcribed spacer 5’ ETS (RNAPI transcript) involves both channel threading and channel-independent pathways to access Rrp44.
(A) Northern analysis of RNAs coprecipitated with immunoaffinity purified (IP) active Rrp44-HTP (WT), Rrp44-exo-HTP (exo) or Rrp44-exo-S1-HTP (exo-S1), along with 2% input RNA. RNA species are detected with a probe hybridizing near the TSS of the 5’ ETS (+49–67, see panel B for location of the probe). Sybr safe staining for 5S rRNA is shown as loading control. (B) Distribution of reads across the 5’ ETS, recovered with Mtr4, Rrp6, Csl4, Rrp41 in an Rrp44-exo background, and Rrp44-exo and Rrp44-exo-S1, normalized to millions of mapped reads. Scale is linear. A diagram of the 5’ ETS and the 18S rRNA is also shown. (C) Model for 5’ ETS degradation. Following cotranscriptional cleavage of the pre-rRNA, the 5’ ETS is oligo-adenylated by TRAMP and targeted to Rrp44 through the channel. The 5’ ETS is subsequently released from the channel (possibly aided by Mtr4 activity) and subjected to new oligo-adenylation by TRAMP, before being targeted to Rrp44 through direct access.