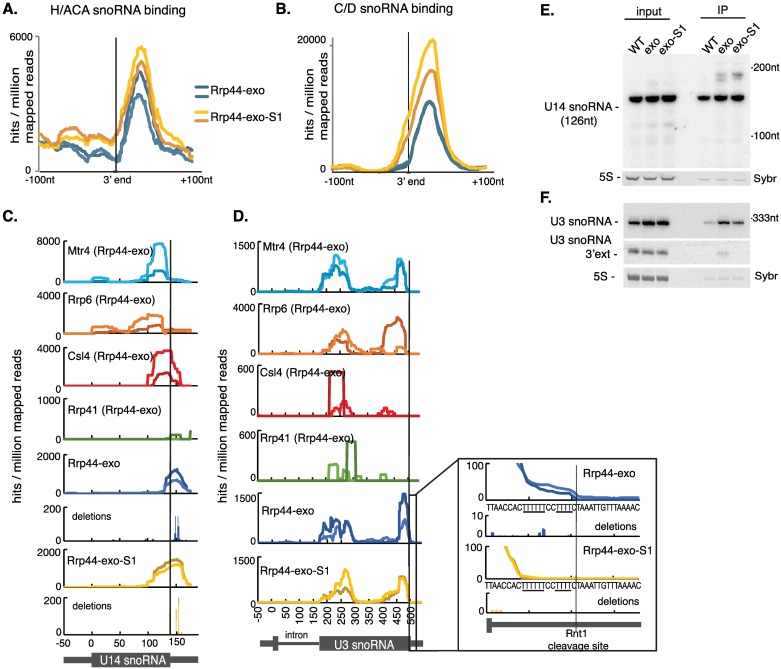
Fig 6. snoRNAs use both channel threading and direct access to Rrp44 for processing.
(A, B) Metagene analyses of all box H/ACA snoRNAs (A) or all box C/D snoRNAs (B) aligned by the 3’ end of the mature snoRNA region. Two independent experiments for Rrp44-exo (blue) and Rrp44-exo-S1 (yellow) are shown. (C, D): Distribution of reads across the box C/D snoRNAs U14 (C) and U3 (D), recovered with Mtr4, Rrp6, Csl4, Rrp41 in the Rrp44-exo background, and Rrp44-exo and Rrp44-exo-S1, normalized by millions of mapped reads. Scale is linear. (E-F) Northern analysis of RNAs coprecipitated with wild type Rrp44-HTP (WT), Rrp44-exo-HTP (exo) or Rrp44-exo-S1-HTP (exo-S1), along with 2% (E) or 1% (F) input RNA, probed for the box C/D snoRNAs U14 (E) or U3 (F). Sybr safe staining for 5S rRNA is shown as a loading control.