Abstract
Purpose
Acute bouts of resistance exercise and subsequent training alters protein turnover in skeletal muscle. The mechanisms responsible for the changes in basal post-absorptive protein turnover and its impact on muscle hypertrophy following resistance exercise training are unknown. To determine whether post-absorptive muscle protein turnover following 12 weeks of resistance exercise training (RET) plays a role in muscle hypertrophy. In addition, we were interested in determining potential molecular mechanisms responsible for altering post-training muscle protein turnover.
Methods
Healthy young men (n=31) participated in supervised whole body progressive RET at 60-80% 1 repetition maximum (1-RM), 3d/wk for 3 months. Pre- and post-training vastus lateralis muscle biopsies and blood samples taken during an infusion of 13C6 and 15N phenylalanine and were used to assess skeletal muscle protein turnover in the post-absorptive state. Lean body mass (LBM), muscle strength (determined by dynamometry), vastus lateralis muscle thickness (MT), myofiber type-specific cross-sectional area (CSA), and mRNA were assessed pre- and post-RET.
Results
RET increased strength (12-40%), LBM (∼5%), MT (∼15%) and myofiber CSA (∼20%) (p<0.05). Muscle protein synthesis (MPS) increased 24% while muscle protein breakdown (MPB) decreased 21% respectively. These changes in protein turnover resulted in an improved net muscle protein balance in the basal state following RET. Further, the change in basal MPS is positively associated (r=0.555, p=0.003) with the change in muscle thickness.
Conclusion
Post-absorptive muscle protein turnover is associated with muscle hypertrophy during resistance exercise training.
Keywords: Skeletal muscle, growth, strength training, mTORC1, ribosome biogenesis
Introduction
Muscle protein turnover (i.e., the concurrent cellular processes of synthesis and breakdown) is stimulated following resistance exercise (RE) (Phillips et al. 1997; Fry et al. 2011). The mechanisms regulating the acute changes in muscle-protein synthesis (MPS) following RE have been well examined (Burd et al. 2009), yet less is understood regarding the adaptation (Wilkinson et al. 2008; Burd et al. 2009) of muscle protein turnover and molecular events (Wilkinson et al. 2008; Leger et al. 2006) (Vissing et al. 2013; Rahbek et al. 2014; Hulmi et al. 2009b) occurring with chronic resistance exercise training (RET). In addition, the influence of acute post-exercise muscle-protein turnover in regulation of muscle hypertrophy has recently been questioned (Mitchell et al. 2014) shifting interest toward other methodological approaches and time frames during the process of muscle hypertrophy. Several studies that have found chronic RET modulates muscle-protein turnover (Phillips et al. 1999; Phillips et al. 2002; Kim et al. 2005; Tang et al. 2008; Wilkinson et al. 2008). In particular, after a period of RET, individuals experience a shorter duration of the acute elevation of MPS following a bout of RE (Damas et al. 2015), and a higher rate of basal post-absorptive MPS compared to pre-training levels (Damas et al. 2015). Therefore, the adaptation of muscle protein turnover throughout chronic RET may be an important contributor to muscle hypertrophy as more recent studies are starting to reveal (Brook et al. 2015; Damas et al. 2016). However, the role of muscle protein turnover, specifically in the basal post-absorptive state, during resistance exercise-induced muscle hypertrophy is unknown.
To better understand the phenotypical relevance of increased post-absorptive MPS following RET, we examined the relationship between the change in protein turnover (pre- to post-training) and muscle hypertrophy in a large cohort (n=31) of young men. In addition, to gain more insight into the mechanisms behind these changes, we comprehensively examined skeletal muscle protein concentration, RNA and DNA concentrations, mechanistic target of rapamycin complex 1 (mTORC1) signaling (Leger et al. 2006; Vissing et al. 2013) and amino acid sensing (Moro et al. 2016) and mRNA expression of several key markers of muscle growth and protein turnover. We hypothesized that resistance exercise training improves the efficiency of post-absorptive muscle protein turnover and is highly associated with the increase in muscle hypertrophy.
Methods
Participants
We recruited healthy male participants for this clinical trial. The participants were recruited through locally posted flyers, newspaper advertisements, and by word of mouth. After initial contact, prospective participants filled out a pre-screening questionnaire to determine eligibility and availability to participate. Potential participants were screened in the morning after an overnight fast at the Institute for Translational Sciences-Clinical Research Center (ITS-CRC) at the University of Texas Medical Branch (UTMB). The screening day included: 3-day food diary analysis, strength testing, a medical history and physical exam, resting electrocardiogram (ECG), and laboratory tests (complete blood count with differential, liver and kidney function tests, coagulation profile, fasting blood glucose, hepatitis B and C screening, an HIV test, thyroid stimulating hormone level, lipid profile, urinalysis, and drug screening). Participants with clinical signs of malnutrition, those who were on anabolic steroids or corticosteroids in the past 6 months, current tobacco users, admitted vegan or vegetarians, individuals on a high-protein diet (>1.6g·kg-1·d-1), high dairy diet (> 6 servings of dairy per day), and those using protein supplements or having dairy allergies were excluded. The participants were healthy and recreationally active but were not engaged in any regular exercise-training program (<2 sessions of high-intensity aerobic or resistance exercise/week) at the time of enrollment. All participants gave written informed consent before enrollment in the study. The study was approved by the UTMB Institutional Review Board, and is in compliance with the Declaration of Helsinki as revised in 1983. Of the 36 participants who underwent baseline testing, 3 withdrew before undergoing exercise training, 1 withdrew during the first 6 weeks and 1 withdrew during the final week. Suitable muscle histological samples were only available for 28 of 31 completers.
Study design
Following enrollment, participants completed a 10-14 day pre-training run-in period that consisted of the pre-training study day at UTMB and then 3 non-consecutive days of exercise familiarization and baseline 1-repetition max (1-RM) strength testing at the University of Texas Medical Branch Alumni Fieldhouse. At the run-in, participants were given a study binder containing study information and food diary record instructions.
The pre-training study day included the stable isotope infusion and muscle biopsies for the assessment of vastus lateralis myofiber size and muscle protein turnover. The day also included assessment of body composition, anthropometry, knee extensor muscle thickness, blood and serum collection, and strength testing via dynamometry. Two to three days later, the participants reported to the UTMB Alumni Fieldhouse for familiarization/testing before beginning 12 weeks of training. After 12 weeks of training, participants were re-tested (post-testing study day) exactly 3 days following the final exercise session of the training program to avoid the acute effects from the final exercise session (Damas et al. 2015). Laboratory tests (complete blood count with differential, liver and kidney function tests, coagulation profile, fasting blood glucose, lipid profile and urinalysis) were conducted by the UTMB clinical laboratory. For the post-testing, participants reported to the ITS-CRC at the same time in the morning as for the pre-training study day to repeat the same laboratory tests and sample collection.
Pre and post-testing study day
Participants reported to the ITS-CRC at UTMB in the morning following an overnight fast. They were instructed to refrain from any medication affecting muscle metabolism, and also caffeine, fish oil supplements, and alcohol for three days before testing. They were instructed to avoid strenuous or long-duration exercise for 3 days before arrival and to drink a liter of water the night before. After arrival on the unit, participants voided to ensure an empty bladder and bowel, and then laid supine for 30 min prior to assessment of body composition by DXA scan (dual-energy X-ray absorptiometry) (Hologic ADR 4500W, Bedford, MA). The same technician set the regions of interest for all the DXA scans.
To maintain a supine position throughout the study day, participants were transported to and from the CRC bed in a stretcher. After the DXA scan, a forearm catheter was placed for tracer infusion, and a retrograde catheter was placed in the opposite hand for arterialized blood sampling. After collecting a background blood sample, a primed-continuous infusion of L-[ring-13C6]phenylalanine (priming dose: 2 μmol/kg; infusion rate: 0.05 μmol/kg/min) and L-[15N]phenylalanine (priming dose: 2 μmol/kg; infusion rate: 0.05 μmol/kg/min) was started to measure basal post-absorptive muscle protein synthesis and breakdown using the precursor-product method (infusion study schematic seen in ESM Figure 1 in the Online Resource #1). At 2 h into the infusion, a percutaneous biopsy sample of the vastus lateralis (VL) muscle was performed using sterile procedures with a 5 mm Bergström biopsy needle (Bergstrom 1962) with suction under local anesthesia (1% lidocaine) to measure the muscle protein synthesis and breakdown rates, gene expression, RNA concentration (as a proxy for ribosomal abundance), muscle water, muscle protein concentration, and cell morphology. A leg was randomly selected for the biopsies and the same leg was biopsied after exercise training. At 4 h into the infusion, the L-[15N]phenylalanine infusion was stopped to begin the measurement of tracer dilution from endogenous tracee released from muscle-protein breakdown. Between 4-5 h, frequent arterialized blood samples were taken from the hand vein to measure phenylalanine enrichments and blood amino acid (AA) concentrations. At the end of the infusion (5h), a second muscle biopsy was taken from the same incision site but sampled at a different angle to measure muscle protein synthesis and breakdown. The biopsy samples were aliquoted, snap-frozen in liquid nitrogen, and stored at -80°C for future analysis. When available, approximately 20 mg of muscle was oriented and embedded in Tissue Tek optimal cutting temperature (OCT; Thermo Fisher Scientific, Rockford, IL) on a cork and frozen in liquid nitrogen-cooled isopentane. Suitable muscle histological samples were only available for 28 of 31 completers. The samples were stored at -80°C for subsequent immunohistochemical analysis.
After the DXA scan, ultrasound (Phillips HDI 5000) of the VL muscle was conducted while the participants remained supine, as previously described (Porter et al. 2015). Briefly, several B-Mode real-time images of the VL were taken in the mid-sagittal position at 50% (mid-belly) and 25% (distal) of the femur length (from the anterior superior iliac spine to the superior border of the patella). The ultrasound head position, pre- and post-training, was placed relative to specific measured landmarks. The image that offered the sharpest contrast with the femur was chosen to ensure perpendicular placement of the scan head. VL muscle thickness was assessed as the average distance from the superficial aponeurosis to the deep aponeurosis at these two locations. Preliminary testing on the same individuals revealed that the coefficient of variation for measurements taken the day of or several weeks apart was 1.42±0.20 and 1.84±0.40%, respectively Because the CV of this measurement is excellent, we used this measure to test the association of muscle protein synthesis and muscle hypertrophy.
Peak torque and power of the knee extensors and knee flexors of the non-biopsied leg were subsequently determined by dynamometry (Biodex Medical, Shirley, NY), as previously conducted (Porter et al. 2015).
Following the strength test, participants were fed a meal before leaving the unit. All testing was repeated on the post-testing day in the same order.
Resistance-exercise training
Following familiarization and 1-repetition maximum (1-RM) strength testing, participants began a 12-week whole-body progressive resistance exercise-training (RET) program. These were pre-training sessions whereby the participants were familiarized to the exercise equipment and had their 1st strength test for assessment of exercise intensity. The 1st familiarization session consisted of a short warm-up and a brief sub-max test to help guide the 1-RM strength testing on the 3rd familiarization session. The second familiarization session consisted of light exercise (2 sets of 10 reps) on each of the exercises. All exercise-training sessions were performed at the UTMB Alumni Fieldhouse. Exercise sessions were performed on non-consecutive days, 3 times weekly, with 4 rest days per week under the supervision of qualified personal trainers. Participants were allowed to maintain their recreational physical activity but instructed not to do any other strength training outside the study. RET was performed at an intensity of 60-80% of 1-RM strength and consisted of 3-4 sets of 8-10 repetitions performed to failure for each exercise, as previously conducted (Porter et al. 2015). 1-RM strength was directly tested on the chest press, leg press, and knee extension. 1-RM strength was estimated with 8-RM strength testing on the remaining exercises. To allow for unforeseen life events, participants were given 13 weeks following the familiarization period to complete 36 exercise sessions on non-consecutive days, typically three times per week. This allowed for 100% exercise compliance.
Nutritional Intake
Participants were instructed to maintain their habitual diet and to log a 3-day food diary on 3 occasions: pre-testing, six weeks, and post-testing. On each occasion, participants were given detailed instructions and were told to record their normal diet in the week before the testing day on two weekdays and one weekend day, with emphasis that one of the days be the day before testing. These records were entered into Nutrition Data System for Research 2012 to estimate energy intake and macro-nutrient composition.
Muscle-Protein Turnover and Whole Body Proteolysis
Blood and tissue amino acid concentrations were determined with isotopic internal standards. Enrichments of free L-[ring-13C6]phenylalanine, L-[15N]phenylalanine, (L-[ring-13C9], L-[15N] phenylalanine), L-[5,5,5-2H3] leucine, [13C] valine and [13C] isoleucine in blood, muscle tissue fluid (MTF) and bound protein (mixed muscle and myofibrillar protein) were measured by gas chromatography-mass spectrometry (GCMS) after addition of appropriate internal standards and precipitation of blood and tissue proteins with sulfosalycilic acid, extraction with cation exchange chromatography, and tert-butyldimethylsilyl derivatization (t-BDMS). Correction for skewed isotopomer distribution and overlapping spectra was performed, as previously described (Wolfe and Chinkes 2005). The incorporation of L-[ring-13C6]phenylalanine in the mixed muscle proteins and myofibrillar protein was measured after protein extraction and hydrolysis, amino acid extraction with cation exchange chromatography, t-BDMS derivatization, and GCMS analysis (Wolfe and Chinkes 2005). Myofibrillar protein extraction for assessment of isotope enrichment was conducted as we have previously described and validated (Reidy et al. 2014c). The fractional synthesis rate (FSR) of mixed muscle proteins was calculated from the incorporation rate of L-[ring-13C6]phenylalanine into the mixed muscle proteins or myofibrillar proteins, and the free-tissue phenylalanine enrichment:
where ΔEP/t is the slope of the straight line that fits the protein-bound phenylalanine enrichment across two sequential biopsies, t is the time interval encompassing the two biopsies, EM(1), and EM(2) are the phenylalanine enrichments (tracer/tracee) in the free muscle pool in the two biopsies. The results are presented as %·h-1. Fractional breakdown rate (FBR) of muscle proteins was measured with multiple phenylalanine tracers using the precursor-product method (Wolfe and Chinkes 2005). The method requires measurement of intracellular free phenylalanine enrichment at steady-state and after 1 h of tracer decay along with frequent arterialized blood sampling. To measure FBR, the ratio of the L-[ring-13C6] and 15N-phenylalanine MTF enrichment at biopsy 1 was used to approximate the 15N-phenylalanine MTF enrichment plateau enrichment at 4 hours using the L-[ring-13C6]-phenylalanine MTF enrichment at 5 hours. The L-[15N] phenylalanine blood enrichments taken at 4-5 hours was used for the 1 h decay enrichments. FBR was calculated using the formula:
where EA and EM are the arterialized and muscle tissue free enrichments, p = EM/(EA-EM) at plateau, and QM/T is the ratio of free to bound phenylalanine in muscle. Whole body proteolysis was determined at both pre- and post-training by the rate of phenylalanine appearance into the blood as infusion rate (0.05 μmol·kg-1·min-1) divided by the average blood 13C6 phenylalanine enrichment (Wolfe and Chinkes 2005).
Measurement of Intracellular Signaling Proteins
To assay common muscle growth and breakdown pathways, phosphorylation of Akt (Thr308 & Thr473), mTOR (Ser2448) 4E-BP1 (Thr37/46), eEF2 (Thr56) and GSK-3α/β (Ser9) and total protein for Akt, mTOR, 4E-BP1, p70S6K1, S6, eEF2, GSK-3α/β, LAMP1, ATG1, LC3B, Beclin-1 and alpha-tubulin was measured using western blot techniques as previously described (Dreyer et al. 2006; Reidy et al. 2014c) with all data expressed relative to a-tubulin and the internal control with phosphorylated proteins also set relative to their total proteins. All antibodies were purchased through Cell Signaling (Danvers, MA). We examined these markers as they have been suggested to be involved with cell growth and muscle hypertrophy following RET (Wilkinson et al. 2008; Leger et al. 2006) (Walker et al. 2013; Vissing et al. 2013; Rahbek et al. 2014; Hulmi et al. 2009b; Bar-Peled et al. 2013; Stefanetti et al. 2014; Hulmi et al. 2009a).
Measurement of Muscle Protein Concentration, DNA Concentration and Water Content
The assay of muscle protein concentration was adapted from the following protocol (Reidy et al. 2014b). Muscle wet weight (∼10 mg) was determined on a precision microbalance, and subsequently the muscle sample was freeze-dried for 48h. Muscle water content was calculated from the difference in dry and weight wet for each muscle sample and expressed as percentage of initial wet weight. The sample was homogenized in 40 volumes of cold buffer (250 mM sucrose, 100 mM potassium chloride, 20 mM imidazole, and 5 mM EDTA; pH 6.8) in a ground glass homogenizer. Samples were centrifuged at 21,000 g for 30 min at 4°C. The supernatant represented the sarcoplasmic protein fraction, and the pellet, representing the myofibrillar protein fraction, was resuspended in 40 volumes of buffer via sonication. Aliquots of the homogenate (total protein), and sarcoplasmic, and myofibrillar protein fractions were measured for protein concentration with the Pierce™ BCA Protein Assay Kit (ThermoFisher, Waltham, MA). The amount of protein in each of the three fractions was normalized to the wet and dry weight of each muscle sample. Muscle water content was determined, as previously described (Reidy et al. 2014b). The total muscle homogenate DNA concentration was determined with a fluorometric assay utilizing the DNA-specific fluorescent Hoechst 33258 dye, according to the methods of Adams and Haddad (Adams and Haddad 1996).
RNA extraction and semiquantitative real-time PCR
RNA isolation, cDNA synthesis, and real-time qPCR were performed as previously described (Porter et al. 2015). Total RNA was isolated by homogenizing 10-20 mg of tissue with a hand-held homogenizing dispenser (T10 Basic Ultra Turrax, IKA, Wilmington, NC) in 1 ml of TRI Reagent®. The RNA was separated into an aqueous phase using 0.2 ml of chloroform and subsequently precipitated from ∼475μl of the aqueous phase using 0.5 ml of isopropanol. Total RNA was quantified by measuring the total volume of the aqueous phase, as previously described (Bickel et al. 2005). This was conducted by calculating the muscle total RNA concentration based on total RNA yield and the weight of the analyzed sample. RNA was washed twice with 1 ml of 75% ethanol, air-dried, and suspended in a known amount of nuclease-free water. RNA concentration was determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Also, RNA concentration, integrity and 18S and 28S ribosomal subunit peak area were assessed with the Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA). The average RNA integrity number was 8.06±0.06 and the 28S-to-18S ribosomal subunit ratio was 1.27±0.02. RNA was DNase-treated using a commercially available kit (DNA-free, Ambion, Austin, TX). A total of 2 μg of RNA was reverse transcribed into cDNA according to the directions provided by the manufacturer (iScript, BioRad, Hercules, CA). Real-time qPCR was carried out using the PrimePCR system with a CFX Connect PCR cycler (BioRad). cDNA was analyzed with SYBR green fluorescence (iQ SYBR green supermix; BioRad). Validated gene targets and unique Bio-Rad assay ID's for the following targets (CDK2, FBX032, FOXO3, MSTN, MYOD1, MYOG, TRIM63, (aka MuRF-1), BNIP3L, CDKN1A, HSPA8, IGF1, LAMP1, LAMP2, MITF, MTOR, PAIP2B, hVps34 (aka PIK3C3), RPLP0, RPS6KB1, TFE3, TFEB, ULK1, URB2) can be seen in Online Resource #1. Also, markers of notch signaling (mib2) and ribosomal biogenesis (pre-rRNA 45S (ETS) were assayed (Online Resource 1). The geometric mean of 3 targets (ACTB, B2M, and RPL13A; Online Resource 1) were originally selected as normalization/housekeeping genes. B2M and ACTB were not stable pre- to post-training; however RLP13A was stable and was utilized as the normalization/housekeeping gene. Muscle samples from 6 participants did not meet target stability criteria required to endure valid analysis. Relative fold changes were determined from the cycle threshold (Ct) values using the 2−ΔΔCt method (Livak and Schmittgen 2001). To simplify data interpretation, the change from pre- to post-training was calculated and presented.
Immunohistochemistry
Samples were removed from the cork at -25°C in a ThermoFisher Cryostat (Fisher Scientific HM 525X) where they were cut in 7 μm cross-sections. Pre- and post-training samples for the same subject were placed on the same slides (Fisherbrand Superfrost®/Plus microscope slides, Fisher Scientific, Wilmington, DE, USA). Immunohistochemical techniques for myosin heavy chain (MHC) type and cross-sectional area (CSA) were conducted as previously described (Fry et al. 2014). Approximately 250 muscle fibers were analyzed for fiber type distribution and 200 for CSA in each sample.
Statistical analysis
All values are expressed as Mean ± SEM or 95% confidence intervals (CI). Data variance and normality were tested and transformations were made, where appropriate. Independent paired t-tests were used to test pre- to post-training differences and each individual time point for blood 15N phenylalanine enrichments. Pearson product correlations were utilized to test associations. Steady state of blood 13C6 phenylalanine enrichments pre- and post-training were examined with linear regression to test for differences in slopes. A 2-way ANOVA, with repeated measures of time and RET was used to test for differences in 15N phenylalanine enrichment and the Bonferroni correction was used to test multiple comparisons when appropriate. Significance was set at p < 0.05 with trends at p 0.05 < 0.1. All calculations were made and figures created in Graph Pad Prizm.6.0f for Mac (La Jolla, CA).
Results
Baseline Characteristics and Body Composition
Descriptive characteristics for all participants at baseline are shown as follows (means with 95% CI) in Table 1.
Table 1. Baseline characteristics1.
| Age, years | 24.8 (23.6,26.1) |
| Height, cm | 177.4 (175.1,179.7) |
| Weight, kg | 76.4 (73.5,79.3) |
| BMI, kg/m2 | 24.3 (23.4,25.2) |
| DXA body composition, kg | |
| Whole body LM | 55.6 (53.2,57.9) |
| Leg LM | 19.1 (18.2,20.0) |
| Fat, % | 23.7 (21.6,25.8) |
| Fat mass | 17.4 (15.6,19.2) |
| 1-Repetition maximum, kg | |
| Average | 80.0 (74.0,85.4) |
| Knee extension | 108.7 (99.5,118.0) |
| Muscle thickness, cm | |
| Knee extensor | 4.18 (3.94,4.41) |
Data are mean ± 95%CI.
LM, lean mass, n=36.
Nutritional Intake
For all participants the average nutritional intakes of energy, carbohydrate, and fat was unchanged during RET and thus pooled as 2361 (2142,2580) kcals, 3.50 (3.20, 3.81) g·kg-1·d-1 and, 1.23 (1.08,1.38) g·kg-1·d-1, respectively. This was, 45.0 (43.1,46.9) and 35.5 (33.9,37.1) % of energy from carbohydrate, and fat, respectively. Protein intake was 1.34 (1.21,1.48), 1.63 (1.38,1.88) and 1.56 (1.33,1.79) g·kg-1·d-1 at pre-, mid-, and post-training, respectively. Protein intake was 18.2 (16.7, 19.8), 20.1 (18.4, 21.9) and 20.2 (18.5, 21.9) % of energy at pre-, mid-, and post-training, respectively. Protein intake increased at mid- (p<0.05), but not post-training and was 1.47 (1.32,1.63) g·kg-1·d-1 on average throughout the training. There was no relationship between protein intake and hypertrophy (p>0.5).
Blood/Cell Counts, Plasma Volume and Metabolic Panel
Complete blood count, electrolytes, urinalysis, and the metabolic lab panel were all within normal limits (ESM Table 1 in Online Resource #2). The exercise training resulted in a 2.2% and 3.9% increase in blood volume and plasma volume, respectively, as calculated via the Dill and Costill method (Costill and Fink 1974), to correct all post values by the plasma volume change (ESM Table 2 in Online Resource #2). There was no change (p<0.05) in cell volume, total carbon dioxide, anion gap, total bilirubin, creatinine, total protein, albumin, alanine aminotransferase, and aspartate aminotransferase following RET. However, hematocrit decreased (p<0.05) and hemoglobin, electrolytes (sodium, potassium, chloride and calcium), glucose, blood urea nitrogen, and alkaline phosphatase increased (p<0.05) following RET.
Amino Acid Concentrations and Muscle Q & T Pool Sizes
Blood phenylalanine, leucine, valine and the sum of all branched-chain amino acids (nmol/ml) increased (p<0.05) following RET, but isoleucine remained unchanged (p>0.10) (ESM Table 3 in Online Resource #2). Muscle intracellular phenylalanine, isoleucine, valine, and the sum of all branched-chain amino acids (nmol/ml) increased (p<0.05) following RET, but intracellular leucine and muscle protein bound phenylalanine remained unchanged (p>0.10) (Fig. 1). The pool size (umol/mg wet tissue) of free muscle phenylalanine (Q) increased (p<0.05) from pre- (65.8±2.0) to post- (77.2±2.1) RET. The pool size (umol/mg wet tissue) of protein-bound muscle phenylalanine (T) remained unchanged (p>0.10) from pre- (44055±1064) to post- (44968±1045) RET. Thus the ratio of T/Q decreased (p<0.05) from pre- (691.5±25.7) to post- (597.8±24.3) RET.
Fig. 1.
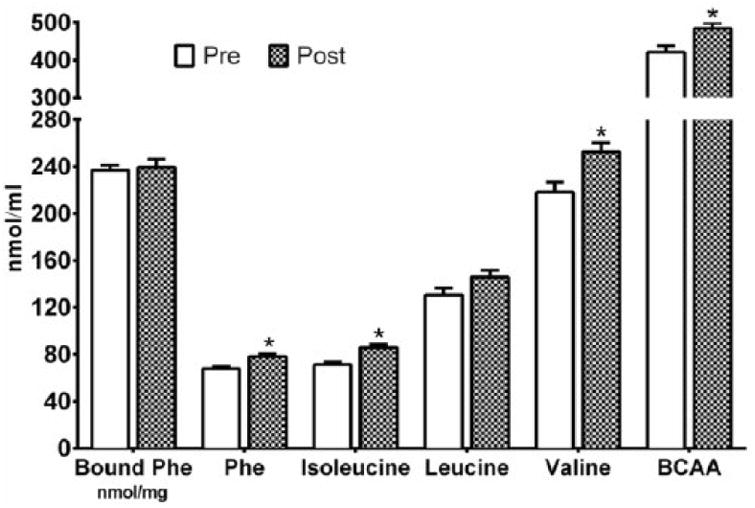
Vastus lateralis free and bound protein phenylalanine (phe), and free isoleucine, leucine, valine, and the sum of all branched-chain amino acid (BCAA) concentrations in young adult men pre- and post-resistance exercise training. * p<0.05 vs Pre.
Myosin Heavy Chain Composition
MHC I composition did not change (p>0.10) following RET, but MHC IIa frequency increased (p<0.05) while hybrid frequency decreased (p<0.05) (data not shown).
Muscle Hypertrophy
Thigh circumference and leg volume increased (p<0.05) following RET (data not shown). Leg, arm, appendicular and whole body lean mass (LM) increased (p<0.05) ∼4-6% following RET (ESM Fig. 2). Ultrasound assessed muscle thickness of both the vastus lateralis (VL) and vastus intermedius (VI) increased (p<0.05) following RET. Vastus lateralis (VL) muscle thickness was 2.26 (2.13,2.38) cm at pre-training and increased 0.32 (0.25,0.38) cm to 2.60 (2.48,2.70) cm post-training (Fig. 3). Vastus intermedius (VI) muscle thickness was 1.93 (1.79,2.07) cm at pre-training and increased 0.27 (0.16,0.38) cm to 2.22 (2.05,2.38) cm post-training. The sum of vastus lateralis (VL) and vastus intermedius (VI) was 4.18 (3.94,4.41) cm at pre-training and increased 0.60 (0.44,0.75) cm to 4.78 (4.57,5.00) cm (p<0.05) following RET (Fig. 2). MHC I, IIa, and hybrid myofiber CSA increased (p<0.05) following RET (Table 2 and ESM Fig. 2).
Fig. 3.
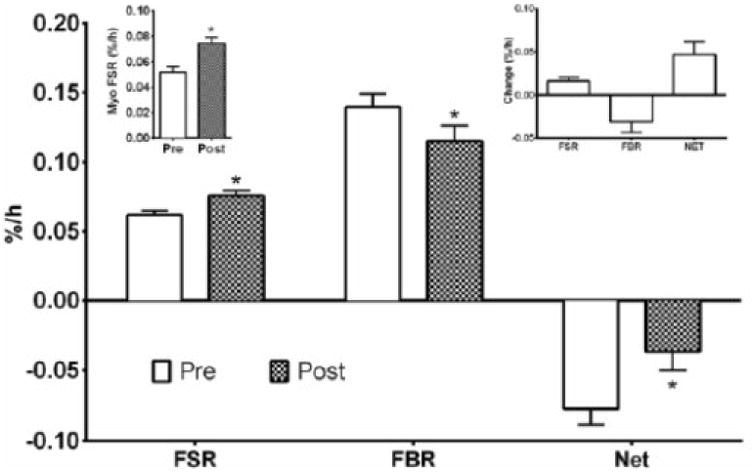
Vastus lateralis muscle protein turnover of young adult men as the absolute values pre- and post-resistance exercise training. The top right inset shows the change in mixed-muscle protein turnover pre- and post-resistance exercise training. The top left inset shows the myofibrillar (Myo) protein synthesis pre- and post-resistance exercise training. Net balance was calculated as the difference in in mixed-muscle protein synthesis and breakdown. FSR, fractional synthesis rate; FBR, fractional breakdown rate, NET, net balance. * p<0.05 vs Pre.
Fig. 2.
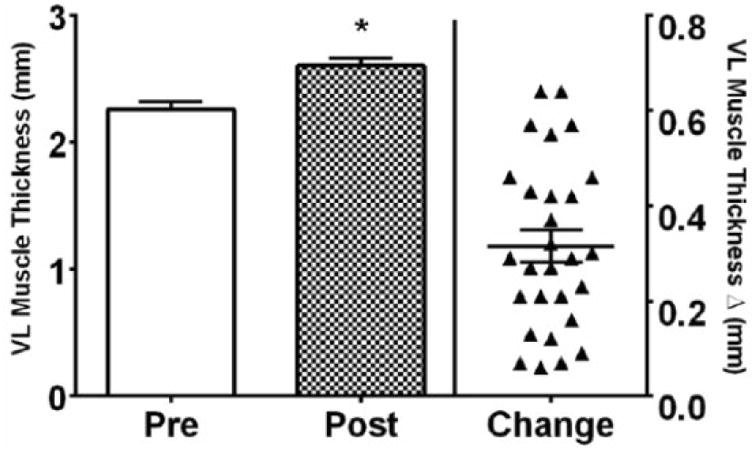
Vastus lateralis (VL) muscle thickness as a measure of muscle hypertrophy in young adult men following resistance exercise training (RET). Absolute values are shown in the left y-axis and change values are in the right y-axis. * p<0.05 vs pre.
Table 2. Pre, Post and absolute change for vastus vateralis myosin heavy chain (MHC) fiber type, myofiber CSA following resistance-exercise training1.
| MHC Typing (relative frequency) | Myofiber CSA | |||||
|---|---|---|---|---|---|---|
| Pre | Post | Change | Pre | Post | Change | |
| I | 32.1±2.4 | 32.0±1.9 | -0.1 (-3.8,3.6) | 4286±188 | 4860±180 | 551 (240,863) |
| IIa | 37.9±1.9 | 44.2±2.7 | 6.3 (1.9,10.8) | 5304±209 | 6220±220 | 923 (578,1268) |
| Hybrids | 30.0±2.0 | 23.8±2.4 | -6.1 (-10.9,-1.4) | 4633±234 | 5663±210 | 1030 (634,1425) |
| All | - | - | - | 4875±203 | 5674±190 | 789 (447,1130) |
Data are Mean and 95% CI.
Boldface, p<0.05 compared to 0. CSA, cross-sectional area.
Dynamometry and 1-RM strength
Isometric torque (Newton-meters) increased (p<0.05) by 33.6 (19.3,47.9) for knee extension and by 14.3 (6.4,22.2) for knee flexion following exercise training. Isokinetic torque (Newton-meters) also increased (p<0.05) by 11.2 (2.9,19.4) for knee extension and by 11.4 (2.0,20.8) for knee flexion following exercise training. For the exercise–training specific exercises, 1-RM strength increased (p<0.05). Baseline knee extension strength was 108.7 (99.5,118.0) kg and increased by 62 (54.9,69.4) kg. The average strength of all 10 exercises was 80.0 (74.0,85.4) kg at baseline and increased by 42.6 (39.5,45.7) kg.
Cell signaling and protein expression
Phosphorylation of Akt (Thr308 & Thr473), mTOR (Ser2448) 4E-BP1 (Thr37/46), eEF2 (Thr56) and GSK-3α/β (Ser9) and total protein for Akt, mTOR, 4E-BP1, p70S6K1, S6, eEF2, GSK-3α/β, LAMP1, ATG1, LC3B, Beclin-1 did not change (p>0.10) following RET (data not shown).
Muscle protein, DNA, RNA concentration and Water and Ribosomal Content
Vastus lateralis RNA concentration increased (p<0.05) following RET (ESM Table 4 in Online Resource #2). The change in RNA concentration positively correlated with the increase in average vastus lateralis thickness (r=0.522, p=0.004) and leg volume (r=0.433, p=0.031). The area of 18S and 28S ribosomal subunits increased (p<0.05) following RET (ESM Table 4 in Online Resource #2). Vastus lateralis muscle protein and DNA concentrations did not change (p>0.10), yet muscle-water content increased (p<0.05) following RET. There was a trend (p<0.10) for the RNA:DNA ratio to increase following RET (ESM Table 4 in Online Resource #2).
Enrichments
Linear regression revealed no change (p>0.05) in the slope of blood 13C6 phenylalanine enrichments at pre-0.041 (-0.009,0.091) and post-training 0.040 (-0.011,0.090) over the course of each experiment. The slopes were equal (p=0.979). Thus, a steady state was present for blood 13C6 phenylalanine. Muscle tissue fluid 13C6 phenylalanine enrichments exhibited a negligible (<0.7%), but significant main effect of time (p<0.001) from the first to second biopsy, within each infusion trial. There was a main effect of RET (p=0.003) demonstrated as a reduction post-training, but not an interaction (p=0.537). Blood 15N phenylalanine enrichment decreased following termination of the tracer infusion as a main effect of time (p<0.01). There was no main effect of RET (p=0.528) or interaction (p=0.104). Muscle tissue fluid 15N phenylalanine enrichments decreased following termination of tracer infusion as a main effect of time (p<0.01). There was no main effect of RET (p=0.198), but a strong trend for an interaction (p=0.054). This was driven by higher muscle tissue fluid 15N phenylalanine enrichment (p<0.05) determined in the second biopsy following RET. ESM Fig. 3 in Online Resource #2 depicts the blood and muscle tissue fluid enrichments.
Muscle protein turnover
MPS, also referred to as the fractional synthesis rate (FSR) was increased (p<0.05) following RET (Fig. 3). When mixed-muscle FSR was calculated with blood enrichments as the precursor, a similar pattern was demonstrated with a 20% increase (p<0.05) from pre- (0.041±0.002%·h-1) to post-training (0.049±0.002%·h-1). MPB, also referred to as fractional breakdown rate (FBR), was decreased (p<0.05) following RET. Thus net protein turnover was less negative (p<0.05). Muscle myofibrillar FSR was increased (∼39%) (p<0.05) following RET (Fig. 3). The change in FSR pre- to post-training was positively associated (r=0.555, p=0.003) with the change in average vastus lateralis muscle thickness (Fig. 4).
Fig. 4.
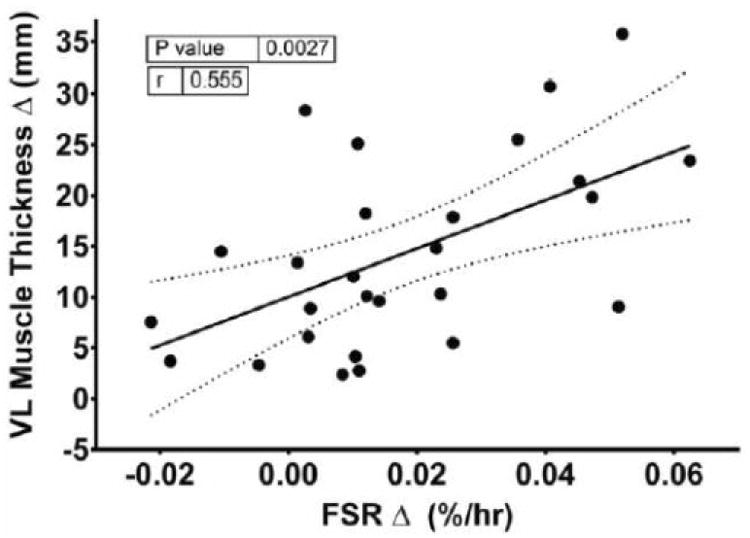
Association between the change in mixed-muscle protein synthesis and muscle thickness in the vastus lateralis (VL) of young adult men following resistance exercise training. FSR, fractional synthesis rate.
Whole body proteolysis
Whole body proteolysis (mmol·-1kg-1·min-1), as assessed via the phenylalanine rate of appearance into blood, increased (p<0.05) from 0.72 (0.69,0.75) to 0.79 (0.75,0.83) following RET.
Muscle growth and turnover mRNA expression
CDK2, MyoG, FBX032, MuRF-1, BNIP3L, CDKN1A, GAPARAP, HSPA8, LAMP1, LAMP3, MITF, MTOR, pre-rRNA 45S (ETS), RPS6KB1, DGZ, TFE3, TP53, ULK1, URB2 and ACTB mRNA expression did not change following RET (data not shown). IGF-1, FOXO3, mib2, MSTN, RPLP0, SLC38A9 and B2M were down-regulated (p<0.05), whereas LAMP2, PAIP2B, hVps34 and TFEB were up-regulated (p<0.05) following RET. There was a trend (p<0.10) for MyoD1 to be down-regulated following RET (Fig. 5).
Fig. 5.
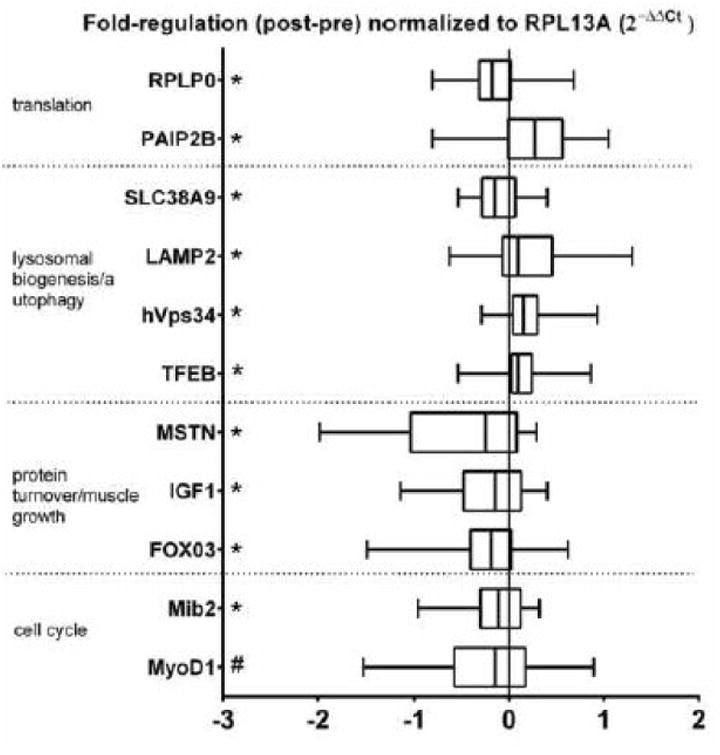
Fold regulation in vastus lateralis mRNA expression of young adult men following resistance exercise training. * p<0.05 vs Pre.
Discussion
In this study we report resistance exercise training (RET)-induced skeletal muscle hypertrophy with increases in lean mass (∼5%), knee extensor muscle thickness (∼15%), myofiber cross-sectional area (∼20%), and leg strength (∼12% via dynamometry). These phenotypic changes were characterized by increased skeletal muscle translational capacity (i.e., total muscle RNA, 18S, and 28S ribosomal subunits). Most notably, post-absorptive protein turnover had a 24% increase in muscle protein synthesis and a 21% decrease in muscle protein breakdown— which resulted in a post-training net balance that was less negative compared to pre-training. Intriguingly, the increase in protein synthesis was positively correlated (r=0.555, p=0.003; Fig. 4) with the change in vastus lateralis muscle thickness following RET. This finding suggests an important role for post-absorptive protein turnover in the modulation of the muscle proteome and as a result of RET, muscle hypertrophy.
The decrease in FBR in our study was mathematically driven by an increase in the product, muscle-tissue free phenylalanine (Q), relative to the precursor content of bound phenylalanine (T) (Figure 1). This resulted in less dilution of muscle-tissue free 15N phenylalanine enrichments from endogenous tracee following RET. Overall, RET induced an expansion of both the blood (phenylalanine, leucine, valine, and total branched- chain) and muscle-tissue free (phenylalanine, isoleucine, valine and total branched- chain) amino acids (AA) suggesting that exercise will expand these AA pools. These increases in tissue-free AA concentration occurred regardless of increased plasma volume (∼4%) and decreased muscle water content (∼1%). The reason for the expanded tissue-free AA pool is unknown and intriguing, but this data coupled with the increased whole-body proteolysis following RET and data from the metabolic panel may represent increased metabolism and protein turnover resulting in post-absorptive AA release in other non-muscle tissues (Waterlow et al. 1978; Cynober 2002), particularly those with higher turnover rates (Tessari et al. 1996) during or following RET. Typically, the muscle would provide AAs to other organs in the post-absorptive state, yet, following RET there could be improved efficiency in protein metabolism to maintain peripheral lean mass. This is an interesting hypothesis that warrants further investigation.
Our relatively larger cohort, for an exercise-training protein metabolism study, was advantageous to better assess the role of post-absorptive protein turnover and muscle growth following RET. Thus, we were able to demonstrate a significant positive association between the change in post-absorptive muscle protein synthesis and muscle hypertrophy (e.g., vastus lateralis muscle thickness) following RET. Throughout the exercise training, protein turnover in the post-absorptive state could be a time frame that characterizes high responders to RET through maintenance of protein mass and/or a limiting of muscle loss in the fasted state. Although our study design is unable to define the precise role and time course of the post-absorptive state during RET on muscle hypertrophy, it is clear that this period is important for muscle hypertrophy as several elegant reports have demonstrated (Brook et al. 2015; Damas et al. 2016). Until recently, the hours following exercise have received the most focus regarding the accrual of new proteins supporting muscle hypertrophy (Mitchell et al. 2014). It was demonstrated that the cumulative MPS during 0-3 weeks, but not 3-6 weeks, of RET drives the muscle hypertrophic response following RET (Brook et al. 2015). Following this study, another use of day-to-day cumulative MPS demonstrated that the integrated MPS response at 3 and 10 weeks of RET is very strongly correlated to muscle hypertrophy (Damas et al. 2016). This suggests that other time periods, including the post-absorptive state, as we demonstrate here, may play more of a role than previously thought.
Most investigations examining basal MPS following RET are conducted 2-4 days (3 days in the current study) following the last exercise bout as this timeframe is well past the point where acute RET-induced MPS increases return to baseline values (ESM Table 5) (Damas et al. 2015). In support of our findings, the results from other studies indicate that post-absorptive mixed-muscle protein synthesis is elevated after RET (Kim et al. 2005) and a trend was demonstrated for higher values in trained individuals compared to untrained individuals (Phillips et al. 1999). However, studies examining postprandial muscle-protein synthesis (i.e., in subjects who were not fasted) following RET are equivocal (Tang et al. 2008; Phillips et al. 2002). We also demonstrated increased myofibrillar protein synthesis, as shown elsewhere (Wilkinson et al. 2008), but in contrast to an earlier study by the same authors (Kim et al. 2005). The reasons for the discrepancy in myofibrillar protein synthesis are unclear, but may stem from differences in the duration of RET, individual differences in the response in smaller (n<10) cohorts (Tang et al. 2008; Wilkinson et al. 2008), or some other unknown factor.
Not only does our study contain the largest cohort (pre-post completers, n=28) of young adults to complete a pre/post assessment of basal post-absorptive protein turnover following RET, but it also is the first study to assess longitudinally both muscle protein synthesis and breakdown. The only study examining post-absorptive MPS and MPB in the context of RET was a cross-sectional comparison between un-trained and RET-trained individuals. Although Phillips et al. (Phillips et al. 1999) found a trend for MPS to be higher in RET trained individuals compared to untrained individuals, there was no difference between groups in their fractional breakdown rate (FBR). However, in our longitudinal design we demonstrated there was a decrease in FBR (∼21%) following RET. Our muscle protein turnover findings are in contrast to the increased post-absorptive FSR and FBR following aerobic exercise training (AET) (Pikosky et al. 2006), and perhaps this can help explain why RET more frequently results in greater muscle hypertrophy as compared to AET. Only one other longitudinal study has examined both FSR and FBR following RET and that study reported both FSR and FBR were increased post-training (Phillips et al. 2002). However, that study involved both men and women and was conducted during post-prandial conditions thus making results between the two studies difficult to reconcile (Phillips et al. 2002).
In addition to our findings, previous research indicated increased MPS in the post-absorptive state following RET (Kim et al. 2005; Phillips et al. 2002; Phillips et al. 1999; Wilkinson et al. 2008), but as some have mentioned (Phillips et al. 2002), we know very little regarding the cellular and molecular mechanisms for these changes. Work from our laboratory has found that activation of the mechanistic target of rapamycin complex 1 (mTORC1) is required for the contraction (Drummond et al. 2009) and amino acid-induced (Dickinson et al. 2011) stimulation of MPS. Yet, we have also shown that mTORC1 is likely not a prominent regulating factor of post-absorptive MPS (Dickinson et al. 2013). In support of this previous finding, we could not demonstrate changes in mTORC1 signaling or protein content in skeletal muscle, as others have also shown (Rahbek et al. 2014; Wilkinson et al. 2008; Brook et al. 2015). Thus, translational efficiency may not be the regulating factor in the post-absorptive state (Vissing et al. 2013; Rahbek et al. 2014), but rather ribosomal content (i.e., translational capacity) (Figueiredo et al. 2015; Nader et al. 2014; von Walden et al. 2012). We used total muscle RNA as a proxy for translational capacity and demonstrated, similar to a recent reports (Figueiredo et al. 2015; Brook et al. 2015), that the increase in total RNA was also positively associated with muscle hypertrophy (e.g., vastus lateralis muscle thickness) following RET. However, in contrast to recent findings (Figueiredo et al. 2015), we could not demonstrate increased expression of a primary marker of ribosomal biogenesis (pre-rRNA 45S) and found that the expression of several mRNA's involved in ribosomal biogenesis or encoding ribosomal proteins did not change (MTOR, RPS6KB1, RPL13A & URB2) while one (RPLP0) even decreased. Further, the mRNA expression of a translational inhibitor (PAIP2B) (Berlanga et al. 2006) was increased suggesting a mechanism to suppress the expanded translational capacity. However, we also demonstrated increased content of the two most abundant ribosomal subunits (18S and 18S), signifying that enhanced ribosomal biogenesis had indeed occurred and suggesting that our measurements may have been after the peak of transcriptional regulation for ribosomal biogenesis, which appears to occur during the initiation of exercise training (Nader et al. 2014; von Walden et al. 2012). Although others have indicated that global protein synthesis in the post-absorptive state could be increased independently of mTORC1 via AKT/GSK-3/elF2B signaling (Wilkinson et al. 2008; Leger et al. 2006), our data and others (Walker et al. 2013; Vissing et al. 2013; Rahbek et al. 2014) demonstrate no change, whereas another report indicates a decrease (Hulmi et al. 2009b). Thus, it appears that translation initiation (i.e., translational efficiency) is not increased following RET via mTORC1 dependent or independent mechanisms to regulate protein turnover in the basal post-absorptive state. Rather, the increase in ribosomal biogenesis and the resultant increase in translational capacity is likely playing a predominant role in post-absorptive muscle protein synthesis.
Since the lysosome is thought to be a key region for amino acid sensing (Bar-Peled et al. 2012), it might be a regulatory site of protein turnover (Bar-Peled et al. 2013). The improved net balance (decreased MPB and increased MPS) following RET suggests an improvement in amino acid sensing driving enhanced intra-cellular recycling of amino acids from breakdown to be re-synthesized into muscle proteins. Paradoxically, yet reflective of the increased intracellular amino acid concentrations detected post-training, RET resulted in downregulation of the lysosomal amino acid sensor SLC38A9. Although we could not demonstrate an increase in autophagic flux markers such as beclin-1 or LC3B, we found an upregulation of several mRNA's encoding proteins (LAMP2, hVps34) under regulation by the lysosomal biogenesis transcription factor (TFEB), which was also increased (Settembre et al. 2012). TFEB is involved in regulating lysosomal content and amino acid sensing suggesting a role for autophagy during post-absorptive RET-induced muscle hypertrophy. Interestingly, the reduction in expression of FOXO3, a known transcription factor in protein degradation, coincides well with the reduced MPB we demonstrated following RET. In contrast to some previous research, (Leger et al. 2006; Stefanetti et al. 2014), but in agreement with other findings (Stefanetti et al. 2014; Hulmi et al. 2009a), we did not see a change in two potential transcriptional targets of FOXO3, (FBX032 and MURF-1), the two main E3 ubiquitin ligases involved in ubiquitin proteasome-mediated protein degradation. This suggests that some other mechanism of protein degradation is being modulated through FOXO3, possibly autophagy.
In an attempt to study participants in a situation more akin to standard living conditions, and to save on expensive hospital inpatient costs, we had participants walk onto the unit the morning of the tracer study. This, and the lack of dietary standardization, may reflect an inability to tease out differences in our protein-expression data (Vissing et al. 2011). Regardless, as support for our current model, we were able to demonstrate robust changes in skeletal muscle protein turnover and also changes in mRNA expression following RET.
There are a few potential explanations as to why our basal FBR values are higher than in some studies (Fry et al. 2013; Phillips et al. 1999) but not in other studies reported in the literature (Phillips et al. 1997; Gundermann et al. 2014). We recently demonstrated that subjects who walk onto the unit (current study) typically have ∼20% higher MPS values than inpatients (Reidy et al. 2014a). Since FSR and FBR are well coupled (Phillips et al. 1997), it is possible that FBR could also be elevated in our walk-on participants. More likely the reason for our higher values and the variation in FBR measures when compared to FSR measures across the literature are probably due to assumptions and additional parameters in the FBR calculation (Wolfe and Chinkes 2005).
In conclusion, we found an improved post-absorptive net muscle protein balance (i.e., increased muscle protein synthesis and reduced muscle protein breakdown) following RET-induced muscle hypertrophy in a large cohort of young men. We provide evidence of a novel role for post-absorptive muscle protein turnover in the regulation of muscle hypertrophy by describing a positive association between the increase in muscle size and the change in MPS following RET. The training induced increase in post-absorptive MPS appears to be the result of enhanced translational capacity (i.e., increased ribosomal biogenesis) rather than improved translational efficiency. The reduction in post-training MPB is associated with a reduced expression of FOXO3 and increased expression of genes regulating lysosomal biogenesis/autophagy. The combination of an increase in ribosome and lysosomal biogenesis may be suggestive of improved intracellular amino acid recycling in human skeletal muscle following resistance exercise training. The various phases periods of adaption of protein turnover during RET are now becoming revealed (Brook et al. 2015; Damas et al. 2016) and future studies are encouraged to examine the this in more detail across various subject populations and with emphasis on amino acid recycling/sensitivity.
Supplementary Material
Supplemental Figure 1. Schematic of the infusion study day, which was conducted in the morning after an overnight fast. The post-training assessment was conducted three days after the last exercise session. AC, antecubital.
ESM Fig. 2 Percent change in muscle hypertrophy of young adult men following resistance exercise training (RET) by measure (anthropometry, dual-x ray absorptiometry, ultrasound muscle thickness and myofiber type-specific cross-sectional area) and location (whole body, appendicular and leg). All measures were p<0.05 vs 0. Cross-sectional area (CSA); vastus intermedius (VI); vastus lateralis (VL); lean mass (LM).
ESM Fig. 3 Pre- and post-training blood & muscle tissue fluid (MTF) enrichments
ESM Table 1 Clinical labs: metabolic panel before (Pre) and 12 weeks (Post) resistance-exercise training
ESM Table 2 Clinical labs: blood and cell count and volume before (Pre) and 12 weeks (Post) resistance-exercise training
ESM Table 3 Blood amino acids
ESM Table 4 Water content, protein, DNA, and RNA concentration of the vastus lateralis muscle before (Pre) 12 weeks (Post) resistance-exercise-training
Supplemental Table 5. Summary of human skeletal MPS & MPB following RET in the fasted and fed states
Acknowledgments
We thank the clinical research staff of the Institute for Translational Sciences-Clinical Research Center at UTMB for assisting in screening and consenting patients and participants and for assisting in data collection. We thank Syed Husaini, MD, for his assistance in subject recruitment, screening and performing muscle biopsies. We also thank, DPT Samantha Dillon, DPT Matthew Nguyen, SPT Benjamin Brightwell, Camille Brightwell and SPT Jennifer Thedinga for their assistance in supervising the exercise training of research participants. We also thank Dr. Marinel M. Ammenheuser for editing the manuscript.
This project was partially supported by a grant from DuPont Nutrition & Health with assistance from NIH R01 AR49877, T32-HD07539, NIDRR H133P110012, P30 AG024832 and in part by a NIH Clinical and Translational Science Award 5UL1TR001439-02 from the National Center for Advancing Translational Sciences.
This was a subset of the trial registered at clinicaltrials.gov as NCT01749189
Author disclosure: The authors declare that this study was partially funded by DuPont Nutrition & Health. Representatives from DuPont Nutrition & Health were not involved with data collection and laboratory analysis.
Abbreviations
- α-Tub
alpha-Tubulin
- 4E-BP1
Eukaryotic initiation factor 4E binding protein 1
- AA
amino acids
- FSR
fractional synthesis rate
- FBR
fractional breakdown rate
- ACTB
Beta-actin
- Akt
Protein kinase B
- AA
aminoacids
- ATG1
AuTophaGy related 1
- B2M
Beta-2-Microglobulin
- Beclin-1
beclin 1, autophagy related
- BNIP3L
BCL2/adenovirus E1B 19 kDa protein-interacting protein 3-like
- BCAA
branched chain amino acids
- CDKN1A
cyclin-dependent kinase inhibitor 1A, aka (p21, Cip1)
- CDK2
Cyclin-dependent kinase 2
- DGKZ
DiacylglycerolKinase, Zeta
- EAA
essential amino acids
- eEF2
Eukaryotic elongation factor 2
- FBX032
F-Box Only Protein 32,aka MAFbx (Muscle Atrophy F-Box Protein and aka (Atrogin-1)
- FBR
fractional breakdown rate
- FOXO3
Forkhead Box O3
- FSR
fractional synthesis rate
- GAPARAP
GABA(A) receptor-associated protein
- GCMS
Gas chromatography-mass spectrometry
- GSK
Glycogen synthase kinase
- HSPA8
heat shock 70kDa protein 8
- IGF-1
Insulin-like Growth Factor-1
- ITS-CRC
Institute for Translational Sciences-Clinical Research Center
- LAMP1
Lysosomal-associated membrane protein 1
- LAMP2
Lysosomal-associated membrane protein 2
- LAMP3
Lysosomal-associated membrane protein 3
- LC3 α/β
Microtubule-associated protein 1 light chain 3 alpha/beta
- mib2
Mindbomb E3 Ubiquitin Protein Ligase 2
- MITF
Microphthalmia-Associated Transcription Factor
- MSTN
myostatin
- MTOR
mechanistic target of rapamycin
- mTORC1
Mammalian target of rapamycin complex 1
- MuRF-1
Muscle RING-finger protein-1
- MyoD1
Myogenic Differentiation 1
- MyoG
Myogenin (MyogenicFactor 4)
- p70S6K1
p70 ribosomal S6 kinase 1
- PAIP2B
Poly(A) Binding Protein Interacting Protein 2B
- PIK3C3
Phosphoinositide-3-Kinase, Class 3 aka (hVps34)
- pre-rRNA 45S (ETS)
Preribosomal RNA 45S(external transcribed spacers)
- RET
resistance exercise training
- RPLP0
Large Ribosomal Protein
- RPS6KB1
Ribosomal Protein S6 Kinase, 70kDa, Polypeptide 1
- S6- rpS6
Ribosomal protein S6
- Ser
Serine
- SLC38A9
member 9 of the solute carrier family 38
- TFE3
transcription Factor Binding To IGHM Enhancer 3
- TFEB
Transcription Factor EB
- Thr
Threonine
- TP53
Tumor Protein P53
- TTR
Tracer to tracee ratio
- ULK1
Unc-51 Like Autophagy Activating Kinase 1
- URB2
URB2 ribosome biogenesis 2 homolog
Footnotes
ESM Figures 1 and 2 and ESM Tables 1-5 are available from the “Online Resource” link in the online posting of the article.
Authors' contributions: B.B.R., P.T.R., and E.V. designed the research; P.T.R., M.M.M., M.S.B., J.M.D, C.S.F., and R.R.D. conducted the research; B.B.R., E.V., P.T.R., C.S.F., M.M.M., R.R.D., M.S.B. and J.M.D, reviewed the manuscript; P.T.R., M.S.B., and B.B.R. analyzed data; and P.T.R. and B.B.R. wrote the manuscript and had primary responsibility for final content.
Conflicts of Interest: P.T. Reidy, M.S. Borack, M.M. Markofski, J.M. Dickinson, Fry, C.S. R.R. Deer, E. Volpi, B.B. Rasmussen have no conflicts of interest.
References
- Adams GR, Haddad F. The relationships among IGF-1, DNA content, and protein accumulation during skeletal muscle hypertrophy. Journal of Applied Physiology. 1996;81(6):2509–2516. doi: 10.1152/jappl.1996.81.6.2509. [DOI] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340(6136):1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150(6):1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom J. Muscle electrolytes in man. Scandinavian journal of medicine & science in sports. 1962;68:1–110. [Google Scholar]
- Berlanga JJ, Baass A, Sonenberg N. Regulation of poly(A) binding protein function in translation: Characterization of the Paip2 homolog, Paip2B. Rna. 2006;12(8):1556–1568. doi: 10.1261/rna.106506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel CS, Slade J, Mahoney E, Haddad F, Dudley GA, Adams GR. Time course of molecular responses of human skeletal muscle to acute bouts of resistance exercise. J Appl Physiol. 2005;98(2):482–488. doi: 10.1152/japplphysiol.00895.2004. [DOI] [PubMed] [Google Scholar]
- Brook MS, Wilkinson DJ, Mitchell WK, Lund JN, Szewczyk NJ, Greenhaff PL, Smith K, Atherton PJ. Skeletal muscle hypertrophy adaptations predominate in the early stages of resistance exercise training, matching deuterium oxide-derived measures of muscle protein synthesis and mechanistic target of rapamycin complex 1 signaling. FASEB J. 2015;29(11):4485–4496. doi: 10.1096/fj.15-273755. [DOI] [PubMed] [Google Scholar]
- Burd NA, Tang JE, Moore DR, Phillips SM. Exercise training and protein metabolism: influences of contraction, protein intake, and sex-based differences. J Appl Physiol. 2009;106(5):1692–1701. doi: 10.1152/japplphysiol.91351.2008. doi:91351.2008[pii]10.1152/japplphysiol.91351.2008. [DOI] [PubMed] [Google Scholar]
- Costill DL, Fink WJ. Plasma volume changes following exercise and thermal dehydration. J Appl Physiol. 1974;37(4):521–525. doi: 10.1152/jappl.1974.37.4.521. [DOI] [PubMed] [Google Scholar]
- Cynober LA. Plasma amino acid levels with a note on membrane transport: characteristics, regulation, and metabolic significance. Nutrition. 2002;18(9):761–766. doi: 10.1016/s0899-9007(02)00780-3. [DOI] [PubMed] [Google Scholar]
- Damas F, Phillips S, Vechin FC, Ugrinowitsch C. A review of resistance training-induced changes in skeletal muscle protein synthesis and their contribution to hypertrophy. Sports medicine. 2015;45(6):801–807. doi: 10.1007/s40279-015-0320-0. [DOI] [PubMed] [Google Scholar]
- Damas F, Phillips SM, Libardi CA, Vechin FC, Lixandrao ME, Jannig PR, Costa LA, Bacurau AV, Snijders T, Parise G, Tricoli V, Roschel H, Ugrinowitsch C. Resistance training-induced changes in integrated myofibrillar protein synthesis are related to hypertrophy only after attenuation of muscle damage. J Physiol. 2016;594(18):5209–5222. doi: 10.1113/JP272472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson JM, Drummond MJ, Fry CS, Gundermann DM, Walker DK, Timmerman KL, Volpi E, Rasmussen BB. Rapamycin does not affect post-absorptive protein metabolism in human skeletal muscle. Metabolism: clinical and experimental. 2013;62(1):144–151. doi: 10.1016/j.metabol.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson JM, Fry CS, Drummond MJ, Gundermann DM, Walker DK, Glynn EL, Timmerman KL, Dhanani S, Volpi E, Rasmussen BB. Mammalian target of rapamycin complex 1 activation is required for the stimulation of human skeletal muscle protein synthesis by essential amino acids. J Nutr. 2011;141(5):856–862. doi: 10.3945/jn.111.139485. doi:jn.111.139485[pii]10.3945/jn.111.139485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer HC, Fujita S, Cadenas JG, Chinkes DL, Volpi E, Rasmussen BB. Resistance exercise increases AMPK activity and reduces 4E-BP1 phosphorylation and protein synthesis in human skeletal muscle. J Physiol. 2006;576(Pt 2):613–624. doi: 10.1113/jphysiol.2006.113175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond MJ, Fry CS, Glynn EL, Dreyer HC, Dhanani S, Timmerman KL, Volpi E, Rasmussen BB. Rapamycin administration in humans blocks the contraction-induced increase in skeletal muscle protein synthesis. J Physiol. 2009;587(Pt 7):1535–1546. doi: 10.1113/jphysiol.2008.163816. doi:jphysiol.2008.163816[pii]10.1113/jphysiol.2008.163816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo VC, Caldow MK, Massie V, Markworth JF, Cameron-Smith D, Blazevich AJ. Ribosome biogenesis adaptation in resistance training-induced human skeletal muscle hypertrophy. American journal of physiology Endocrinology and metabolism. 2015;309(1):E72–83. doi: 10.1152/ajpendo.00050.2015. [DOI] [PubMed] [Google Scholar]
- Fry C, Drummond M, Glynn E, Dickinson J, Gundermann D, Timmerman K, Walker D, Dhanani S, Volpi E, Rasmussen B. Aging impairs contraction-induced human skeletal muscle mTORC1 signaling and protein synthesis. Skeletal Muscle. 2011;1(1):11. doi: 10.1186/2044-5040-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry CS, Drummond MJ, Glynn EL, Dickinson JM, Gundermann DM, Timmerman KL, Walker DK, Volpi E, Rasmussen BB. Skeletal muscle autophagy and protein breakdown following resistance exercise are similar in younger and older adults. J Gerontol A Biol Sci Med Sci. 2013;68(5):599–607. doi: 10.1093/gerona/gls209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry CS, Noehren B, Mula J, Ubele MF, Westgate PM, Kern PA, Peterson CA. Fibre type-specific satellite cell response to aerobic training in sedentary adults. J Physiol. 2014;592(Pt 12):2625–2635. doi: 10.1113/jphysiol.2014.271288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundermann DM, Walker DK, Reidy PT, Borack MS, Dickinson JM, Volpi E, Rasmussen BB. Activation of mTORC1 signaling and protein synthesis in human muscle following blood flow restriction exercise is inhibited by rapamycin. American journal of physiology Endocrinology and metabolism. 2014;306(10):E1198–1204. doi: 10.1152/ajpendo.00600.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulmi JJ, Kovanen V, Selanne H, Kraemer WJ, Hakkinen K, Mero AA. Acute and long-term effects of resistance exercise with or without protein ingestion on muscle hypertrophy and gene expression. Amino acids. 2009a;37(2):297–308. doi: 10.1007/s00726-008-0150-6. [DOI] [PubMed] [Google Scholar]
- Hulmi JJ, Tannerstedt J, Selanne H, Kainulainen H, Kovanen V, Mero AA. Resistance exercise with whey protein ingestion affects mTOR signaling pathway and myostatin in men. J Appl Physiol. 2009b;106(5):1720–1729. doi: 10.1152/japplphysiol.00087.2009. doi:00087.2009[pii]10.1152/japplphysiol.00087.2009. [DOI] [PubMed] [Google Scholar]
- Kim PL, Staron RS, Phillips SM. Fasted-state skeletal muscle protein synthesis after resistance exercise is altered with training. J Physiol. 2005;568(Pt 1):283–290. doi: 10.1113/jphysiol.2005.093708. doi:jphysiol.2005.093708[pii]10.1113/jphysiol.2005.093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger B, Cartoni R, Praz M, Lamon S, Deriaz O, Crettenand A, Gobelet C, Rohmer P, Konzelmann M, Luthi F, Russell AP. Akt signalling through GSK-3beta, mTOR and Foxo1 is involved in human skeletal muscle hypertrophy and atrophy. J Physiol. 2006;576(Pt 3):923–933. doi: 10.1113/jphysiol.2006.116715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mitchell CJ, Churchward-Venne TA, Parise G, Bellamy L, Baker SK, Smith K, Atherton PJ, Phillips SM. Acute post-exercise myofibrillar protein synthesis is not correlated with resistance training-induced muscle hypertrophy in young men. PloS one. 2014;9(2):e89431. doi: 10.1371/journal.pone.0089431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro T, Ebert SM, Adams CM, Rasmussen BB. Amino Acid Sensing in Skeletal Muscle. Trends Endocrinol Metab. 2016;27(11):796–806. doi: 10.1016/j.tem.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader GA, von Walden F, Liu C, Lindvall J, Gutmann L, Pistilli EE, Gordon PM. Resistance exercise training modulates acute gene expression during human skeletal muscle hypertrophy. J Appl Physiol. 2014;116(6):693–702. doi: 10.1152/japplphysiol.01366.2013. [DOI] [PubMed] [Google Scholar]
- Phillips SM, Parise G, Roy BD, Tipton KD, Wolfe RR, Tamopolsky MA. Resistance-training-induced adaptations in skeletal muscle protein turnover in the fed state. Can J Physiol Pharmacol. 2002;80(11):1045–1053. doi: 10.1139/y02-134. [DOI] [PubMed] [Google Scholar]
- Phillips SM, Tipton KD, Aarsland A, Wolf SE, Wolfe RR. Mixed muscle protein synthesis and breakdown after resistance exercise in humans. Am J Physiol. 1997;273(1 Pt 1):E99–107. doi: 10.1152/ajpendo.1997.273.1.E99. [DOI] [PubMed] [Google Scholar]
- Phillips SM, Tipton KD, Ferrando AA, Wolfe RR. Resistance training reduces the acute exercise-induced increase in muscle protein turnover. Am J Physiol. 1999;276(1 Pt 1):E118–124. doi: 10.1152/ajpendo.1999.276.1.E118. [DOI] [PubMed] [Google Scholar]
- Pikosky MA, Gaine PC, Martin WF, Grabarz KC, Ferrando AA, Wolfe RR, Rodriguez NR. Aerobic exercise training increases skeletal muscle protein turnover in healthy adults at rest. J Nutr. 2006;136:379–383. doi: 10.1093/jn/136.2.379. doi:136/2/379[pii] [DOI] [PubMed] [Google Scholar]
- Porter C, Reidy PT, Bhattarai N, Sidossis LS, Rasmussen BB. Resistance Exercise Training Alters Mitochondrial Function in Human Skeletal Muscle. Med Sci Sports Exerc. 2015;47(9):1922–1931. doi: 10.1249/MSS.0000000000000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahbek SK, Farup J, Moller AB, Vendelbo MH, Holm L, Jessen N, Vissing K. Effects of divergent resistance exercise contraction mode and dietary supplementation type on anabolic signalling, muscle protein synthesis and muscle hypertrophy. Amino acids. 2014;46(10):2377–2392. doi: 10.1007/s00726-014-1792-1. [DOI] [PubMed] [Google Scholar]
- Reidy P, Borack M, Markofski M, Dickinson J, Drummond M, Fry C, Gundermann D, Walker D, Volpi E, Rasmussen B. Inactivity from one overnight hospital stay reduces basal muscle protein synthesis in young adults (820.15) The FASEB Journal. 2014a;28(1 Supplement) [Google Scholar]
- Reidy PT, Hinkley JM, Trappe TA, Trappe SW, Harber MP. Protein composition of endurance trained human skeletal muscle. International journal of sports medicine. 2014b;35(6):476–481. doi: 10.1055/s-0033-1351334. [DOI] [PubMed] [Google Scholar]
- Reidy PT, Walker DK, Dickinson JM, Gundermann DM, Drummond MJ, Timmerman KL, Cope MB, Mukherjea R, Jennings K, Volpi E, Rasmussen BB. Soy-dairy protein blend and whey protein ingestion after resistance exercise increases amino acid transport and transporter expression in human skeletal muscle. J Appl Physiol (1985) 2014c;116(11):1353–1364. doi: 10.1152/japplphysiol.01093.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. The EMBO journal. 2012;31(5):1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanetti RJ, Lamon S, Rahbek SK, Farup J, Zacharewicz E, Wallace MA, Vendelbo MH, Russell AP, Vissing K. Influence of divergent exercise contraction mode and whey protein supplementation on atrogin-1, MuRF1, and FOXO1/3A in human skeletal muscle. J Appl Physiol (1985) 2014;116(11):1491–1502. doi: 10.1152/japplphysiol.00136.2013. [DOI] [PubMed] [Google Scholar]
- Tang JE, Perco JG, Moore DR, Wilkinson SB, Phillips SM. Resistance training alters the response of fed state mixed muscle protein synthesis in young men. Am J Physiol Regul Integr Comp Physiol. 2008;294(1):R172–178. doi: 10.1152/ajpregu.00636.2007. doi:00636.2007[pii]10.1152/ajpregu.00636.2007. [DOI] [PubMed] [Google Scholar]
- Tessari P, Garibotto G, Inchiostro S, Robaudo C, Saffioti S, Vettore M, Zanetti M, Russo R, Deferrari G. Kidney, splanchnic, and leg protein turnover in humans. Insight from leucine and phenylalanine kinetics. The Journal of clinical investigation. 1996;98(6):1481–1492. doi: 10.1172/JCI118937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissing K, McGee S, Farup J, Kjolhede T, Vendelbo M, Jessen N. Differentiated mTOR but not AMPK signaling after strength vs endurance exercise in training-accustomed individuals. Scandinavian journal of medicine & science in sports. 2013;23(3):355–366. doi: 10.1111/j.1600-0838.2011.01395.x. [DOI] [PubMed] [Google Scholar]
- Vissing K, McGee SL, Farup J, Kjolhede T, Vendelbo MH, Jessen N. Differentiated mTOR but not AMPK signaling after strength vs endurance exercise in training-accustomed individuals. Scandinavian journal of medicine & science in sports. 2011 doi: 10.1111/j.1600-0838.2011.01395.x. [DOI] [PubMed] [Google Scholar]
- von Walden F, Casagrande V, Ostlund Farrants AK, Nader GA. Mechanical loading induces the expression of a Pol I regulon at the onset of skeletal muscle hypertrophy. American journal of physiology Cell physiology. 2012;302(10):C1523–1530. doi: 10.1152/ajpcell.00460.2011. [DOI] [PubMed] [Google Scholar]
- Walker S, Hulmi JJ, Wernbom M, Nyman K, Kraemer WJ, Ahtiainen JP, Hakkinen K. Variable resistance training promotes greater fatigue resistance but not hypertrophy versus constant resistance training. European journal of applied physiology. 2013;113(9):2233–2244. doi: 10.1007/s00421-013-2653-4. [DOI] [PubMed] [Google Scholar]
- Waterlow JC, Garlick PJ, Millward DJ. Protein turnover in mammalian tissues and in the whole body. North-Holland Pub. Co. ; sole distributors for the U.S.A. and Canada, Elsevier North-Holland, Amsterdam; New York, New York: 1978. [Google Scholar]
- Wilkinson SB, Phillips SM, Atherton PJ, Patel R, Yarasheski KE, Tarnopolsky MA, Rennie MJ. Differential effects of resistance and endurance exercise in the fed state on signalling molecule phosphorylation and protein synthesis in human muscle. J Physiol. 2008;586(Pt 15):3701–3717. doi: 10.1113/jphysiol.2008.153916. doi:jphysiol.2008.153916[pii]10.1113/jphysiol.2008.153916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe RR, Chinkes DL. Isotope tracers in metabolic research : principles and practice of kinetic analysis. 2nd. Wiley-Liss; Hoboken, N.J: 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Schematic of the infusion study day, which was conducted in the morning after an overnight fast. The post-training assessment was conducted three days after the last exercise session. AC, antecubital.
ESM Fig. 2 Percent change in muscle hypertrophy of young adult men following resistance exercise training (RET) by measure (anthropometry, dual-x ray absorptiometry, ultrasound muscle thickness and myofiber type-specific cross-sectional area) and location (whole body, appendicular and leg). All measures were p<0.05 vs 0. Cross-sectional area (CSA); vastus intermedius (VI); vastus lateralis (VL); lean mass (LM).
ESM Fig. 3 Pre- and post-training blood & muscle tissue fluid (MTF) enrichments
ESM Table 1 Clinical labs: metabolic panel before (Pre) and 12 weeks (Post) resistance-exercise training
ESM Table 2 Clinical labs: blood and cell count and volume before (Pre) and 12 weeks (Post) resistance-exercise training
ESM Table 3 Blood amino acids
ESM Table 4 Water content, protein, DNA, and RNA concentration of the vastus lateralis muscle before (Pre) 12 weeks (Post) resistance-exercise-training
Supplemental Table 5. Summary of human skeletal MPS & MPB following RET in the fasted and fed states


