Abstract
Peptide triazole (PT) antagonists interact with gp120 subunits of HIV-1 Env trimers to block host cell receptor interactions, trigger gp120 shedding, irreversibly inactivate virus and inhibit infection. Despite these enticing functions, understanding the structural mechanism of PT-Env trimer encounter has been limited. In this work, we combined competition interaction analysis and computational simulation to demonstrate PT binding to the recombinant soluble trimer, BG505 SOSIP.664, a stable variant that resembles native virus spikes in binding to CD4 receptor as well as known conformationally-dependent Env antibodies. Binding specificity and computational modeling fit with encounter through complementary PT pharmacophore Ile-triazolePro-Trp interaction with a 2-subsite cavity in the Env gp120 subunit of SOSIP trimer similar to that in monomeric gp120. These findings argue that PTs are able to recognize and bind a closed prefusion state of Env trimer upon HIV-1 encounter. The results provide a structural model of how PTs exert their function on virion trimeric spike protein and a platform to inform future antagonist design.
Keywords: HIV-1, Env trimer, peptide triazole, SOSIP, virus inactivation
Introduction
Human Immunodeficiency Virus Type 1 (HIV-1) is the causative agent of AIDS, which has affected millions of people worldwide. In host cell infection, the HIV-1 envelope (Env) trimeric complex of glycoproteins gp120 and gp41 mediates virus entry by interaction with host cell receptors.1 Sequential receptor binding of gp120 to CD4 and a chemokine receptor (usually CXCR4/CCR5) causes conformational changes in Env leading to gp41 exposure, cell membrane interaction, and fusion of virus and host cell membranes.1,2 As the only virus-specific protein on the surface of the virion, the Env trimeric complex is a crucial viral target to block the cascade of binding and structural rearrangements that lead to cell entry and infection.3
Peptide triazoles (PTs) have been identified, inspired by an initially reported peptide from phage display,4 as a class of HIV-1 entry inhibitors that (1) bind Env gp120 from a wide range of virus subtypes with nanomolar affinities, (2) suppress interactions of gp120 with both CD4 and co-receptor and (3) cause shedding of gp120 from the virion and consequent irreversible inactivation of the virus.5–9 Furthermore, macrocyclic peptide triazoles (cPTs) have been discovered that are proteolytically stable and offer candidate leads to therapeutically useful agents.10 In the light of these attractive properties,11 a better understanding of the mechanism of encounter of PTs with trimeric Env complex would provide a path for enhancing structure-based design in order to obtain more potent inhibitors and to derive improved insights of how Env antagonists of the PT class are able to inactivate the virus upon Env protein binding. We therefore investigated binding of PTs to a trimeric Env construct BG505 SOSIP.664.gp140 (SOSIP), a stabilized HIV-1 Env trimeric derivative3,12–15 that has recently been proposed to represent closed prefusion conformational states of the virion spike protein based on binding specificities for different conformationally dependent Env antibodies.16,17,18 This Env trimer was shown to retain functions of native protein, including binding to CD4 and to various conformation-dependent antibodies.3,15 Further, SOSIP emerged as the first stabilized trimer class that is amenable to crystallographic analysis,12 and several atomic-resolution structures have been derived.12,16,19
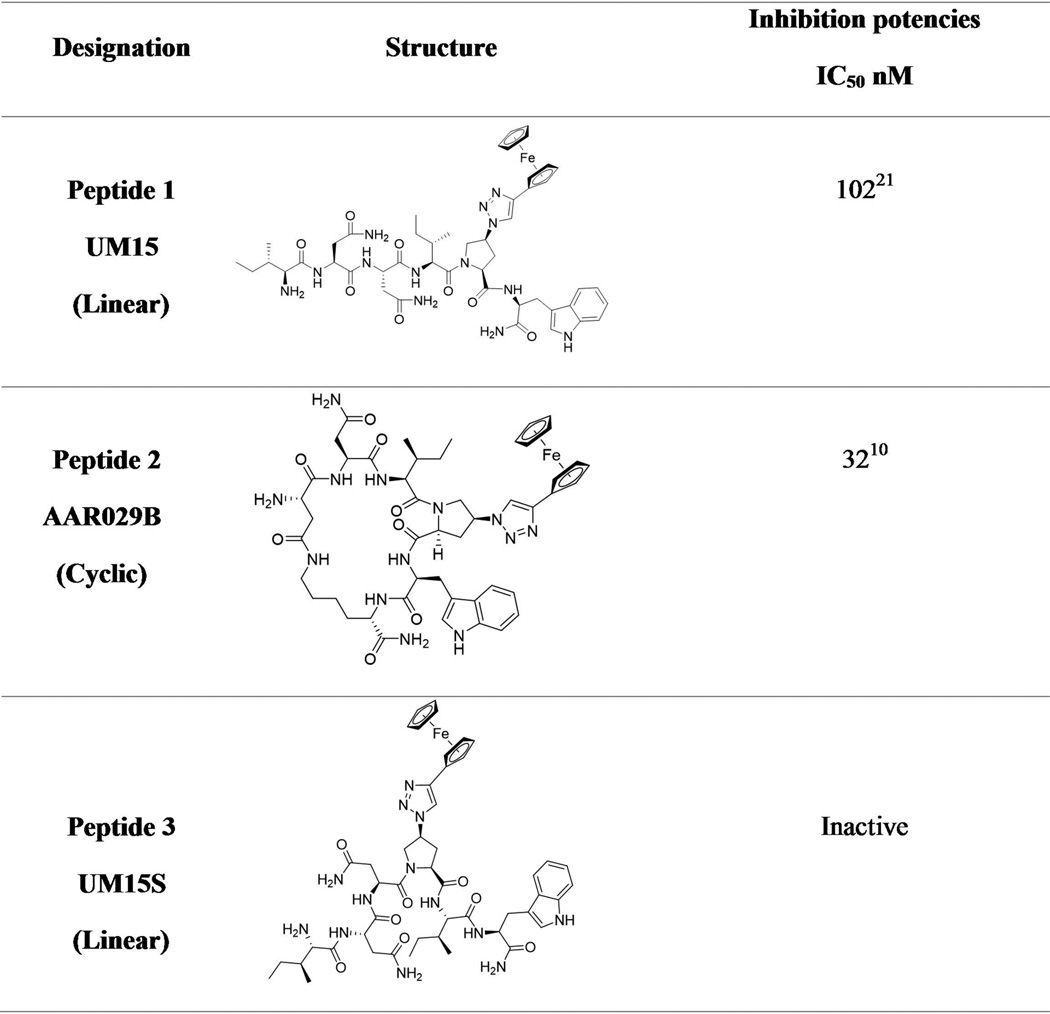
We used both biochemical and computational analyses to investigate the interaction of PTs with SOSIP trimer protein. Peptide triazole derivatives used in this study, namely linear peptide 1 (UM15)20 and cyclic peptide 2 (AAR029b)10, both contain the functional Ile-ferrocenyl-triazoloPro-Trp (I-X-W) pharmacophore.10,20 We found that both peptides 1 and 2 competed with receptor CD4IgG2 for binding to SOSIP proteins and could be modeled into a two-subsite binding cavity, in the trimer protomer, that is similar to that found in gp120 monomer.21 In the light of advancing SOSIP high resolution structures12,16,18,22 and their use for computation-based ligand design23, the results reported here provide a framework for optimization of PT virus inactivators as well as for understanding their structural mechanism of inactivation upon binding to HIV envelope protein.
Materials and Methods
Source of Env Proteins and Antibodies
Recombinant BG505 SOSIP.664.gp140 was expressed in CHO-K1 cells and purified by affinity chromatography using a 2G12 column followed by size exclusion chromatography as described previously.19 CD4IgG2 antibody was obtained from the NIH AIDS Reagent Program, Division of AIDS, and NIAID. Goat anti-Human IgG-Fc Fragment (A80-104A) was purchased from Bethyl Laboratories, Inc.
Peptide Synthesis
Peptide triazole derivatives 1 (UM15), 2 (AAR029b) and 3 (UM15S) were synthesized using standard solid phase Fmoc chemistry on a Liberty Blue microwave peptide synthesizer and validated as described previously.10,21
Competition ELISA
The ability of peptides to inhibit binding of BG505 SOSIP.664 to CD4IgG2 was measured using competition ELISA. Recombinant purified BG505 SOSIP.664 (100 ng) was immobilized by direct adsorption onto a 96-well microtiter plate for 2 hrs at 25 °C. The plate was blocked with 3% BSA in 1× PBS for 90 mins at 25 °C. For the CD4IgG2 competition experiments, 1.5 nM of CD4IgG2 in the presence of increasing concentrations of PT derivatives was added to each well for 1 hour at 25 °C. After washing three times with PBST, horseradish peroxidase (HRP) conjugated anti-human antibody was added at a 1:5000 dilution and incubated for 1 hour at 25 °C. The extent of HRP conjugate binding was detected by adding 200 ul of o-phenylenediamine dihydrochloride (Sigma-Aldrich) for 30 min and measuring optical density at a wavelength of 450 nm using a microplate reader (Molecular Devices).
Surface Plasmon Resonance (SPR) Interaction Analysis
SPR experiments were performed on a BIACORE 3000 optical biosensor (GE) at 25 °C using standard PBS buffer containing 0.005% tween 20. Three flow cells in the CM5 chip were used for amine coupling of different ligands through standard 1-ethyl-3-(3-(dimethylamino)propyl) carbodiimide (EDC)/N-hydroxysuccinamide (NHS) chemistry. Flow cell 2, containing ~10,000 RUs of immobilized anti-Fc alone, served as a control for flow cells 1 and 3, both of which contained 800 RUs of CD4IgG2 captured on the same amount of immobilized anti-Fc as in flow cell 2. Initially, we tested direct binding of gp120 to CD4IgG2 in order to establish that the captured CD4IgG2 was functional. The gp120 analyte at 100 nM was injected over flow cells 1, 2 and 3. For regeneration after each sample run, bound gp120 and CD4IgG2 were removed with 10 mM Tris HCl (2 pulses of 10 s), and CD4IgG2 was captured again for the next cycle of experiments. Further, gp120 binding to CD4IgG2 was competed by flowing a mixture of gp120 and UM15 as analyte. This set of experiments also was carried out with immobilized monomeric gp120 protein to validate the assay. Similar experiments to the above were then performed with SOSIP trimers. In brief, CD4IgG2 (800 RU) was captured via immobilized anti-Fc (10,000 RU), and BG505 SOSIP.664 binding was measured in the presence and absence of serially diluted concentrations of peptide 1. Surface regeneration in this case was achieved by injecting two 10 s pulses of 50 mM Tris HCl. All experiments were done in sets of three. Data analysis of SPR competition data was performed using BIA evaluation v4.1.1 software (GE). To correct for nonspecific binding, response signals from buffer injection and from control flow cell were subtracted from all sensorgrams. Inhibition potencies were determined by calculating the inhibitor concentration required for 50% inhibition of maximal binding (IC50). The inhibition curve was plotted and then fitted using the four-parameter equation as shown below using OriginPro 8 graphing software.
where, Rhigh is the response at the highest inhibitor concentration and Rlow at low inhibitor concentration; concn is the concentration of inhibitor and A1 and A2 are fitting parameters.
Molecular Modeling of The Peptide Triazoles with BG505.SOSIP.664 Crystal Structure (PDB 4NCO12)
Peptide preparation for docking
Peptides 1–3 were built using the ligand-building tool of the Schrödinger package (Schrödinger Suite 2014; Schrödinger, LLC). The minimized structures were then saved as pdb files for the docking simulations. Autodock tools24 graphical interface was then used to prepare the peptides for docking.
Macrocyclic peptide 2 conformational sampling for docking
Macrocyclic peptide 2 (AAR029b) conformational sampling was performed by means of 100 ns MD simulation in explicit solvent by means of NAMD 2.10 using the CHARMM 36 force field with the same parameters described in the “MD simulation” section below. The 100 ns trajectory was then clustered on peptide 2 heavy atoms excluding the ferrocene moiety, using the linkage algorithm, setting 0.11 as cutoff, by means of GROMACS 5.0.4. Twelve clusters were obtained, and the most representative structure of each cluster was then individually subjected to flexible docking.
Flexible docking
The Schrödinger package (Schrödinger Suite 2014; Schrödinger, LLC) was used to pretreat the protein structure (chain A from the trimer structure pdb code 4NCO12) with the protein preparation wizard in Maestro 9.9, where the missing side chains were added and refined. The pretreated protein structure was then prepared by Autodock tools graphical interface (MGtools 1.5.6rc3), where non-polar hydrogens were merged, Kollman charges added and Gasteiger charges calculated. Trp112 residue was set as flexible for the docking,24,25 wherein the indole side chain can move during the docking simulation to uncover the hydrophobic pocket in the inner domain gated by this residue, as we previously showed in monomeric gp120.21 The grid box for the docking search was set to 52 × 52 × 52 points for the x, y and z dimension with a spacing grid of 0.375 Å. AutoGrid 4.2 algorithm was used to evaluate the binding energies between the peptide and the protein and to generate the energy maps for the docking run.
MD simulation of selected docked poses
MD simulation was performed by means of NAMD 2.1026 using the CHARMM 36 force field.27 Ferrocenyl parameters compatible with the CHARMM force field were taken from Hatten et al. 2007.28 To confirm ferrocenyl parameters, a 10 ns MD simulation of 1 (UM15) in vacuum was performed. To set up the system for the subsequent MD simulations of 1, 2 and 3 -4NCO complexes, a first minimization in vacuum (only H atoms and then the whole system for a total of 1100 minimization steps) was performed, followed by solvation of the minimized complex in a TIP3P water box 10 Å longer than the protein in all six directions. Five neutralizing Cl− ions were added. During production MD, the temperature was kept constant at 310 °K by coupling all non-hydrogen atoms to a Langevin thermostat with a friction coefficient of 5 ps−1. Non-bonded interactions were cut off above 10 Å and smoothed to zero beginning from 9 Å. PME long range electrostatics with a grid spacing of 2 Å was used, and all bonds involving hydrogen atoms were constrained using RATTLE.29 In order to keep the distance of the cyclopentadienyl rings to the coordinating iron in the ferrocenyl moiety constant, NAMD 2.10 collective variables were used, setting the distance between the iron and each C atom of the two cyclopentadienyl rings at 2.05 Å, and the distance between the two cyclopentadienyl rings at 3.70 Å.28 Production runs were performed at constant pressure and temperature of 1 bar and 300 °K, respectively (NPT ensemble).
MM-GBSA calculation of SOSIP-PTs complexes
In order to estimate the relative binding free energy of the SOSIP-PT complexes, the MM-GBSA (Molecular Mechanics-Generalized Born Surface Area) method was employed. The relative binding energy was estimated on a 50 ns MD trajectory for all of the peptides 1, 2 and 3, with a 10 ps interval, and explicit water and ions removed. The MM-GBSA was performed on three subsets for each system: SOSIP alone, the PT alone, and the complex. For each of these subsets, the average potential energy was extracted over the simulation that was performed using the generalized implicit solvent module of NAMD 2.10, setting the dielectric constant for the solvent at 78.5. The binding free energy for each SOSIP/PT complex was calculated as follows:
where G is the average potential energy over the simulation. The per-residue energy contribution to PT binding to SOSIP was calculated following the same protocol.
Results
Binding Analyses Using ELISA and SPR
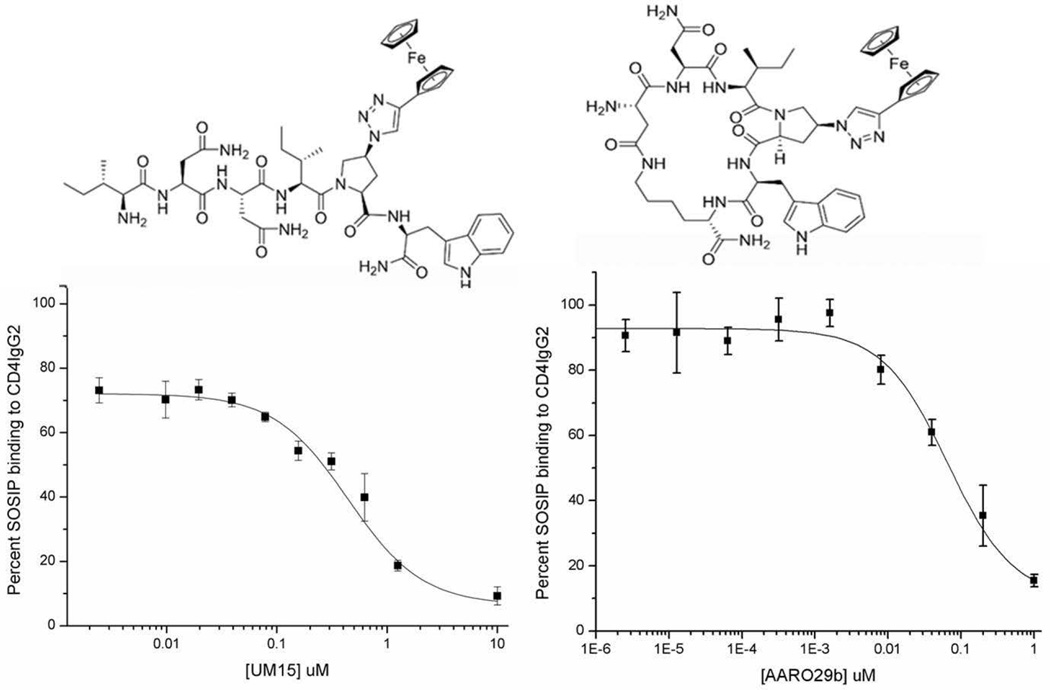
We determined the ability of PTs to bind to BG505 SOSIP.664 using competition ELISA. Two peptide triazole derivatives were evaluated, namely linear peptide 1 (UM15)21 and cyclic peptide 2 (AAR029b)10, both of which contained the functional Ile-ferrocenyltriazoloPro-Trp (I-X-W)20 pharmacophore (Figure 1). Direct binding of CD4IgG2 to plate-immobilized BG505 SOSIP.664 established the assay protocol. Competition of CD4IgG2 binding to the SOSIP protein by both peptides 1 and 2 was shown to be dose-dependent, with IC50 values of 442 nM and 75 nM, respectively (Figure 2).
Figure 1. Peptide denotations, structures and published monomeric gp120 inhibition potencies of peptide triazoles used in this study.
Figure 2. Comparison of peptide 1 (Left) and peptide 2 (Right) structures and competition of CD4IgG2 binding to SOSIP.
Top: Structures of 1 & 2. Bottom: Dose response curves for Peptides 1 and 2 determined from the effect of BG505 SOSIP.664 interaction with CD4IgG2 via ELISA (n=4). The IC50 values of peptides 1 and 2 for CD4IgG2 binding were 442 +/− 2.25nM and 75 +/− 1.2 nM, respectively.
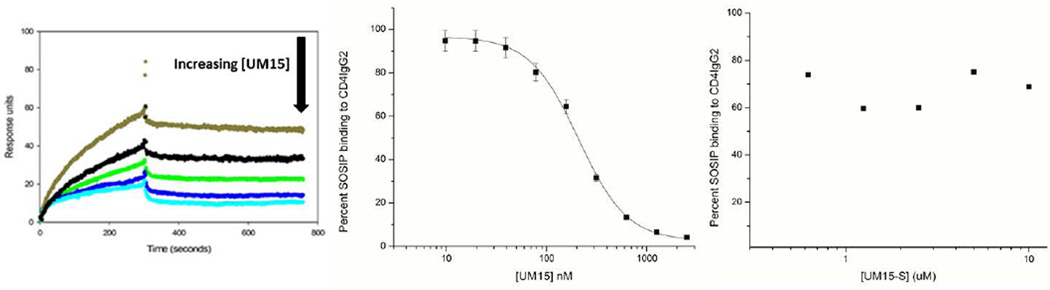
We also used SPR competition assays, in this case to confirm the specificity of PT binding to trimeric SOSIP. In this assay, increasing concentrations (0–200 nM) of SOSIP protein were passed over a surface with medium density (800 RU) CD4IgG2 captured via chip-immobilized anti-Fc, and dose-response results validated this assay. We then compared the effects of peptide 1 and the negative control peptide 3 on SOSIP binding (Figure 3). Dose-dependent inhibition of SOSIP binding to CD4IgG2 was observed by peptide 1 (Figure 3 left and middle), with a mean inhibitory concentration (IC50) of 280 nM. In contrast, no competition was observed with the pharmacophore-scrambled peptide 3 (UM15S, Figure 3 right).
Figure 3. Competition SPR analysis of peptide 1 binding to trimeric BG505 SOSIP.664.gp140.
(Left) Representative sensorgrams showing dose-dependent inhibition of trimeric BG505 SOSIP.664 binding to CD4IgG2 by peptide 1. (Middle) Dose response curve derived from suppression of BG505 SOSIP.664 – CD4IgG2 binding sensorgrams by peptide 1. (Right) Negative control scrambled peptide 3, showing no inhibition of BG505 SOSIP.664 binding to CD4IgG2 (n=2).
Flexible Docking
We previously reported the importance of W112 for linear PT binding to monomeric gp12021 by showing the effect of mutating this residue to alanine. Here, in order to rationalize using the previously applied flexible W112 docking protocol21 for peptide 2, we first evaluated binding of macrocyclic peptide 2 to W112A mutant monomeric gp120 protein. SPR analysis showed dose dependent inhibition of W112A binding to CD4 by peptide 2 with a mean inhibitory concentration of 15 µM. The decreased binding of 2 to W112A gp120 suggests (Figure S1) that macrocyclic peptide 2 also needs W112 side chain for binding, similar to its linear analogues.
Given the observation of specific and high affinity binding of PTs 1 and 2 to SOSIP trimer (Figures 2 and 3), we attempted to develop structural models of the Env binding mode of the PTs. De novo flexible docking (allowing flexible movement of the W112 side chain) was performed, based on earlier docking observations,21 on the gp140 protomer extracted from the crystal structure of BG505 SOSIP.664 (4NCO)12. This structure has recently been validated as a target for investigating gp120-binding ligands.23 For docking the ferrocene containing peptide with the protein in a high accuracy mode, the maximum number of evaluations in Autodock 4.2 (25,000,000) was used. Fifty runs were generated for each peptide by using Autodock 4.2 Lamarckian genetic algorithm24,25 for the searches. Cluster analysis was performed on docking results, with a root-mean-square tolerance of 2.0 Å. For peptide 1, autodock predicted 47 clusters; those with high energies (30 clusters with positive binding energy values) were discarded. The remaining lower energy clusters (negative binding energy values) were examined visually by VIDA 4.2.0 (Openeye Scientific Software, Santa Fe, NM. http://www.eyesopen.com), and pose selection was based on (1) contact with the previously identified protein hot spots21 and (2) structure activity relationship of peptide triazole inhibitors. This resulted in 4 poses that met the selection criteria, and these were then subjected to MD simulations (see below).
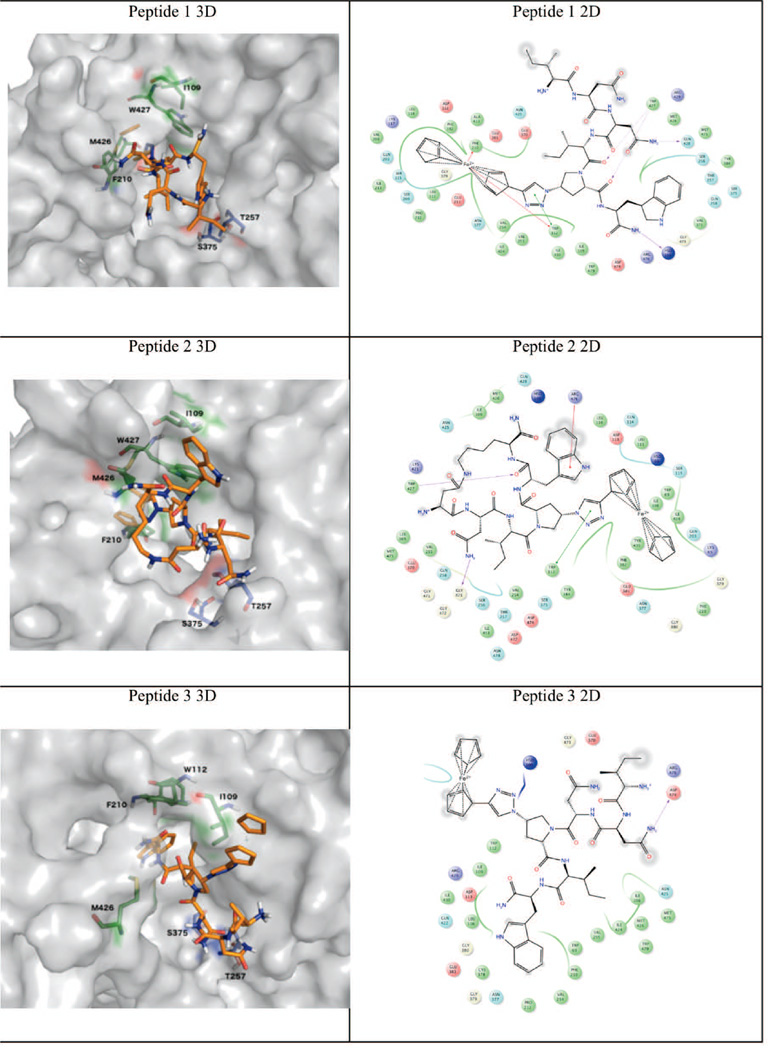
For macrocyclic peptide 2, the MD-generated twelve conformers were individually docked using the same protocol. Conformers returned from docking with unfavorable binding energy (positive values) were excluded. The lowest binding energy pose among the remaining conformers, also matching the mutation data,21 and therefore was selected as a representative binding mode. For peptide 3, low energy poses were not identified, and in this case docking was characterized by altered binding contacts with T257 and S375, residues previously shown by mutational analysis to be important for productive PT binding.21 The docking results suggested that PTs 1 and 2 bind to the SOSIP gp120 subunit through Ile-ferrocenyltriazolePro-Trp pharmacophore interaction at a prototypic two-subsite region21 that overlaps the CD4 binding site.
MD Simulation of The Docked Complexes
Molecular dynamics simulation (50 ns) was used to examine the stability of all PT 2-subsite binding poses with SOSIP coordinates obtained from the flexible docking analysis above. The allowed rotation of W112 χ1 side-chain dihedral angle led to an opening of a small hydrophobic accessory pocket, lined by L108, I109, W112, F210, V255 and M426, from the CD4 binding site. Similar pocket occupancy was previously observed when using the same docking protocol with monomeric gp120 structures.21 The putative poses of 1 and 2 (Figure 4) showed the importance of the indole ring, isobutyl and the ferrocenyl-triazole moieties, of the IXW pharmacophore,20 for protein interactions. Although the linear PT 1 and the constrained cPT 2 may differ in their orientations (indole and isobutyl groups, Figure 4), they still retained contacts with the protein hotspots.21 PT 3, however, was also predicted to form a complex with gp140, a clear false positive. We therefore estimated the binding energy through the MM-GBSA method on each docked complex over 50 ns MD trajectories. As shown in Table 1, the overall MM-GBSA energy for PTs 1 and 2 were favorable and that for PT 3 was strongly unfavorable, which is consistent with the experimental observations shown in Figures 3 and 4. Moreover, the two-subsite region was shown, in Table 1, to contribute the majority of favorable interaction energy to the overall MM-GBSA energies, with strongly favorable interactions at residues I109, W112, F210, and M426 as observed via a per-residue energy decomposition analysis (Table S1). This is consistent with previous experimentally supported models of earlier generation PT binding to core monomeric gp120.
Figure 4. 3D (left column) and 2D (right column) representations of selected docked PT/SOSIP complexes after refinement with MD simulation.
Table 1.
MM-GBSA average energy values for SOSIP complexes with 1, 2 and 3 over their respective 50 ns MD trajectories, and per-residue energy contribution of subsite 121 and subsite 221 residues to peptide binding.
| PT | MM-GBSA average energy kcal mol−1 (50 ns MD) |
Per-residue energy decomposition analysis (kcal mol−1) |
|||||
|---|---|---|---|---|---|---|---|
| Subsite 1 | Subsite 2 | ||||||
| T257 | S375 | I109 | W112 | F210 | M426 | ||
| 1 | −53.53 | 0.55 | −0.90 | −0.54 | −6.84 | −8.26 | −10.21 |
| 2 | −48.05 | −3.49 | −0.94 | −0.94 | −7.56 | −1.07 | −7.37 |
| 3 | 160.41 | 1.06 | 0.12 | −6.29 | −8.11 | −1.62 | −4.87 |
The main difference in binding between active (1 and 2) and inactive (3) PTs can be ascribed to the contribution of subsite 1 residues and I109 in subsite 2.
Discussion
The overall study reported here was initiated to establish an improved experimental path to better define the structural mechanism of peptide triazole encounter with the HIV-1 Env trimer that leads to virus inactivation by triggering gp120 shedding. Recent advancements in the structural analysis of Env trimers have given rise to several atomic resolution crystal structures12,16,22,30 as well as increased-resolution structures determined by cryo-electron microscopy.14,31,32 Both PT 1 and cPT 2 were found to bind specifically and with high affinity to SOSIP as judged by competition of CD4IgG2 binding. The putative binding poses of PTs with Env protein trimer can initiate hypothesis-building for how PTs interfere with bridging sheet formation, that is required for co-receptor binding, and causes irreversible virus inactivation upon gp120 shedding.
We previously found that PTs compete with monoclonal antibody 17b (as a co-receptor surrogate)7,8,20 for binding to gp120 by disrupting bridging sheet formation.33 In prototypic virus cell infection, CD4 binding to gp120 causes large conformational changes, including bridging sheet formation between V1/V2 loop and β20/β21 strand and subsequent release of the V3 loop that provides a binding site for co-receptor,34 after trimer opening. In contrast, PTs disrupt this conformational change cascade and inhibit binding at the co-receptor binding site in addition to CD4 binding inhibition.7,8,10,20,21 Since the SOSIP structure lacks a formed bridging sheet,12 the observed SOSIP binding by PTs argues that a structured bridging sheet domain is not required for their binding to Env trimer. In turn, these results also suggest that PTs exert their inactivation functions on virus by binding to the closed state of Env trimer, which has undissociated V1/V2 and V3 loops at the trimer apex and therefore does not have a formed bridging sheet. The ability to obtain putative binding poses matching the mutational data using the SOSIP coordinates further supports this hypothesis. The SOSIP coordinates (PDB 4NCO12) have been recently used as a productive tool in structure-based design of different gp120 targeting ligands.23
Based on the gp120-gp41 interface in the recently reported SOSIP trimer structure,16 along with the hypothesized SOSIP-PT binding model presented here, we surmise that PT interactions with Env trimer gp120 could well perturb the α1-helix and other inner domain elements of gp120 involved in interactions with gp41 of Env protein.12,14,32,35 In this view, disruption of these layers by PT binding may be an important cause of observed gp120 shedding from the virion triggered by PTs, leading to irreversible virus inactivation. The model presented here provides insights on the proximity of residues in the PT/cPT binding site to residues in the gp120-gp41 interface and disruptions in the latter that could underlie PT-triggered shedding.
The results of this work demonstrate the potential for deepening mechanistic investigations of PT-Env interactions through use of SOSIP trimer. These include: [1] high resolution structural analysis of PTs complexed to SOSIP protein and, through this, visualization of the atomic details of inhibitor/trimer encounter; and [2] visualizing the impact of PT binding on the interface between gp120 and gp41, and follow-up mutational analysis to investigate the structural mechanism of gp120 shedding upon virion interaction with PTs, the function that causes virus inactivation before host cell encounter. A cautionary note in using SOSIP trimer for structural correlations is that this protein was recombinantly engineered to stabilize its conformation, and conformational constraints of benefit for structural analysis could lead to distortions of binding modes that would otherwise occur with naturally occurring Env trimer. Also, the constrained SOSIP structure could limit predictions of how shedding occurs with PTs. Nonetheless, with several conformationally related crystallographic SOSIP structures12,16,22,30 and improving resolution Cryo-EM structures of SOSIP and non-SOSIPs Env trimers14,32 currently emerging, correlating binding of PTs to these trimer forms ultimately will help establish inhibitor-Env models that can be tested by mutagenic effects with trimers on unmodified viruses.
Supplementary Material
Acknowledgments
This work was funded by the National Institutes of Health through P01GM56550-17(IC), R37AI36082 (JPM and PJK) and P01AI110657 (JPM). We thank A. Cupo and M. Golabek for production of the SOSIP protein used in this work. We thank Openeye Scientific Software (Santa Fe, NM. http://www.eyesopen.com) for providing a complimentary academic license of their software package.
Abbreviations
- AIDS
acquired immune deficiency syndrome
- CCR5
chemokine receptor CCR5
- cPT
cyclic peptide triazole
- CXCR4
chemokine receptor CXCR4
- ELISA
enzyme-linked immunosorbent assay
- MM-GBSA
molecular mechanics energies Generalized Born and Surface Area
- PT
peptide triazole
- SPR
Surface Plasmon Resonance
- wt
wild type
References
- 1.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393(6686):705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 2.Doms RW. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology. 2000;276(2):229–237. doi: 10.1006/viro.2000.0612. [DOI] [PubMed] [Google Scholar]
- 3.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. A Next-Generation Cleaved, Soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, Expresses Multiple Epitopes for Broadly Neutralizing but Not Non-Neutralizing Antibodies. Plos Pathog. 2013;9(9):e1003618. doi: 10.1371/journal.ppat.1003618. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrer M, Harrison SC. Peptide ligands to human immunodeficiency virus type 1 gp120 identified from phage display libraries. J Virol. 1999;73(7):5795–5802. doi: 10.1128/jvi.73.7.5795-5802.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastian AR, Contarino M, Bailey LD, Aneja R, Moreira DRM, Freedman K, McFadden K, Duffy C, Emileh A, Leslie G, Jacobson JM, Hoxie JA, Chaiken I. Interactions of peptide triazole thiols with Env gp120 induce irreversible breakdown and inactivation of HIV-1 virions. Retrovirology. 2013;10:153. doi: 10.1186/1742-4690-10-153. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastian AR, Kantharaju, McFadden K, Duffy C, Rajagopal S, Contarino MR, Papazoglou E, Chaiken I. Cell-Free HIV-1 Virucidal Action by Modified Peptide Triazole Inhibitors of Env gp120. Chem med chem. 2011;6(8):1335–1339. doi: 10.1002/cmdc.201100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gopi H, Umashankara M, Pirrone V, LaLonde J, Madani N, Tuzer F, Baxter S, Zentner I, Cocklin S, Jawanda N, Miller SR, Schon A, Klein JC, Freire E, Krebs FC, Smith AB, Sodroski J, Chaiken I. Structural determinants for affinity enhancement of a dual antagonist peptide entry inhibitor of human immunodeficiency virus type-1. Journal of Medicinal Chemistry. 2008;51(9):2638–2647. doi: 10.1021/jm070814r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gopi HN, Tirupula KC, Baxter S, Ajith S, Chaiken IM. Click chemistry on azidoproline: high-affinity dual antagonist for HIV-1 envelope glycoprotein gp120. Chem med chem. 2006;1(1):54–57. doi: 10.1002/cmdc.200500037. [DOI] [PubMed] [Google Scholar]
- 9.McFadden K, Fletcher P, Rossi F, Kantharaju, Umashankara M, Pirrone V, Rajagopal S, Gopi H, Krebs FC, Martin-Garcia J, Shattock RJ, Chaiken I. Antiviral Breadth and Combination Potential of Peptide Triazole HIV-1 Entry Inhibitors. Antimicrob Agents Ch. 2012;56(2):1073–1080. doi: 10.1128/AAC.05555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rashad AA, Kalyana Sundaram RV, Aneja R, Duffy C, Chaiken I. Macrocyclic Envelope Glycoprotein Antagonists that Irreversibly Inactivate HIV-1 before Host Cell Encounter. J Med Chem. 2015;58(18):7603–7608. doi: 10.1021/acs.jmedchem.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaiken I, Rashad AA. Peptide triazole inactivators of HIV-1: how do they work and what is their potential? Future Med Chem. 2015;7(17):2305–2310. doi: 10.4155/fmc.15.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klasse PJ, Depetris RS, Pejchal R, Julien JP, Khayat R, Lee JH, Marozsan AJ, Cupo A, Cocco N, Korzun J, Yasmeen A, Ward AB, Wilson IA, Sanders RW, Moore JP. Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein. J Virol. 2013;87(17):9873–9885. doi: 10.1128/JVI.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yasmeen A, Ringe R, Derking R, Cupo A, Julien JP, Burton DR, Ward AB, Wilson IA, Sanders RW, Moore JP, Klasse PJ. Differential binding of neutralizing and non-neutralizing antibodies to native-like soluble HIV-1 Env trimers, uncleaved Env proteins, and monomeric subunits. Retrovirology. 2014;11:41. doi: 10.1186/1742-4690-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scharf L, Wang H, Gao H, Chen S, McDowall AW, Bjorkman PJ. Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1. Env. Cell. 2015;162(6):1379–1390. doi: 10.1016/j.cell.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong R, Xu K, Zhou TQ, Acharya P, Lemmin T, Liu K, Ozorowski G, Soto C, Taft JD, Bailer RT, Cale EM, Chen L, Choi CW, Chuang GY, Doria-Rose NA, Druz A, Georgiev IS, Gorman J, Huang JH, Joyce MG, Louder MK, Ma XC, Mckee K, O'Dell S, Pancera M, Yang YP, Blanchard SC, Mothes W, Burton DR, Koff WC, Connors M, Ward AB, Kwong PD, Mascola JR. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science. 2016;352(6287):828–833. doi: 10.1126/science.aae0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, Bylund T, Choi CW, Davison JR, Georgiev IS, Joyce MG, Kwon YD, Pancera M, Taft J, Yang Y, Zhang B, Shivatare SS, Shivatare VS, Lee CC, Wu CY, Bewley CA, Burton DR, Koff WC, Connors M, Crispin M, Baxa U, Korber BT, Wong CH, Mascola JR, Kwong PD. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016;165(4):813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A. 2013;110(11):4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umashankara M, McFadden K, Zentner I, Schon A, Rajagopal S, Tuzer F, Kuriakose SA, Contarino M, Lalonde J, Freire E, Chaiken I. The active core in a triazole peptide dual-site antagonist of HIV-1 gp120. Chem med chem. 2010;5(11):1871–1879. doi: 10.1002/cmdc.201000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aneja R, Rashad AA, Li H, Kalyana Sundaram RV, Duffy C, Bailey LD, Chaiken I. Peptide Triazole Inactivators of HIV-1 Utilize a Conserved Two-Cavity Binding Site at the Junction of the Inner and Outer Domains of Env gp120. J Med Chem. 2015;58(9):3843–3858. doi: 10.1021/acs.jmedchem.5b00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garces F, Lee JH, de Val N, de la Pena AT, Kong L, Puchades C, Hua YZ, Stanfield RL, Burton DR, Moore JP, Sanders RW, Ward AB, Wilson IA. Affinity Maturation of a Potent Family of HIV Antibodies Is Primarily Focused on Accommodating or Avoiding Glycans. Immunity. 2015;43(6):1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moraca F, Acharya K, Melillo B, Smith AB, 3rd, Chaiken I, Abrams CF. Computational Evaluation of HIV-1 gp120 Conformations of Soluble Trimeric gp140 Structures as Targets for de Novo Docking of First- and Second-Generation Small-Molecule CD4 Mimics. J Chem Inf Model. 2016;56(10):2069–2079. doi: 10.1021/acs.jcim.6b00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris GM, Goodsell DS, Huey R, Olson AJ. Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J Comput Aided Mol Des. 1996;10(4):293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 26.Darve E, Rodriguez-Gomez D, Pohorille A. Adaptive biasing force method for scalar and vector free energy calculations. J Chem Phys. 2008;128(14):144120. doi: 10.1063/1.2829861. [DOI] [PubMed] [Google Scholar]
- 27.Henin J, Fiorin G, Chipot C, Klein ML. Exploring Multidimensional Free Energy Landscapes Using Time-Dependent Biases on Collective Variables. J Chem Theory Comput. 2010;6(1):35–47. doi: 10.1021/ct9004432. [DOI] [PubMed] [Google Scholar]
- 28.de Hatten X, Cournia Z, Huc I, Smith JC, Metzler-Nolte N. Force-field development and molecular dynamics simulations of ferrocene-peptide conjugates as a scaffold for hydrogenase mimics. Chem-Eur J. 2007;13(29):8139–8152. doi: 10.1002/chem.200700358. [DOI] [PubMed] [Google Scholar]
- 29.Andersen HC. Rattle - a Velocity Version of the Shake Algorithm for Molecular-Dynamics Calculations. J Comput Phys. 1983;52(1):24–34. [Google Scholar]
- 30.Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, Soto C, Terry DS, Yang Y, Zhou T, Ahlsen G, Bailer RT, Chambers M, Chuang GY, Doria-Rose NA, Druz A, Hallen MA, Harned A, Kirys T, Louder MK, O'Dell S, Ofek G, Osawa K, Prabhakaran M, Sastry M, Stewart-Jones GB, Stuckey J, Thomas PV, Tittley T, Williams C, Zhang B, Zhao H, Zhou Z, Donald BR, Lee LK, Zolla-Pazner S, Baxa U, Schon A, Freire E, Shapiro L, Lee KK, Arthos J, Munro JB, Blanchard SC, Mothes W, Binley JM, McDermott AB, Mascola JR, Kwong PD. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1. Env. Nat Struct Mol Biol. 2015;22(7):522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol. 2013;20(12):1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351(6277):1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emileh A, Tuzer F, Yeh H, Umashankara M, Moreira DR, Lalonde JM, Bewley CA, Abrams CF, Chaiken IM. A model of peptide triazole entry inhibitor binding to HIV-1 gp120 and the mechanism of bridging sheet disruption. Biochemistry. 2013;52(13):2245–2261. doi: 10.1021/bi400166b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A. 2010;107(3):1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.