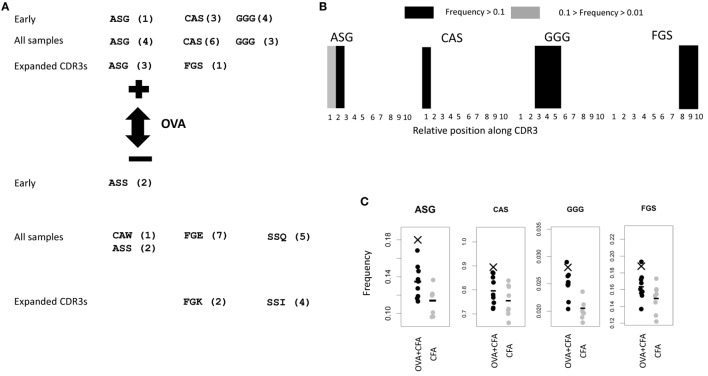
Figure 6.
(A) Common features selected by linear programming boosting (LPBoost) algorithm (with SVM) using singlet CDR3s (those which appear once only) from repertoires from early repertoires only, all repertoires, and expanded CDR3s as defined in Figure 2D. Only those features selected in a proportion of examples above a threshold are shown. The thresholds are shown in Figure S3 in Supplementary Material. The numbers in brackets show the ranking of the triplet in the plots shown in Figure S3 in Supplementary Material. (B) The LPBoost algorithm selects some features with positive coefficients in the equation defining the classifying hyperplane. A higher frequency of these features in a repertoire favors a prediction of an ovalbumin (OVA) + complete Freund’s adjuvant (CFA) classification for that repertoire. By contrast, the LPBoost algorithm selects some features with negative coefficients in the equation defining the classifying hyperplane. A higher frequency of these features in a repertoire favors a prediction of CFA only classification for that repertoire (i.e., it predicts the absence of OVA from the immunization). The panel shows the position of the OVA + CFA predictive triplets along the CDR3. Each CDR3 within a random sample of 106 is divided into 10 equal size units of length (in units of number of amino acids). Individual triplets are assigned a position determined by the relative position of the starting amino acid. The figures show the frequency with which each triplet is found at each position. Black: frequency ≥ 10%; gray: 10% > frequency ≥ 1%; and white: frequency < 1%. (C) The frequency (count per 104 CDR3s) of the CFA + OVA-predictive triplets in total early immunized OVA (black) or CFA (gray) repertoires. Bars show average of each population. X shows the frequency of the triplet in the subset of CDR3s which are found at high frequency in OVA repertoires which are illustrated in Figure 3A.