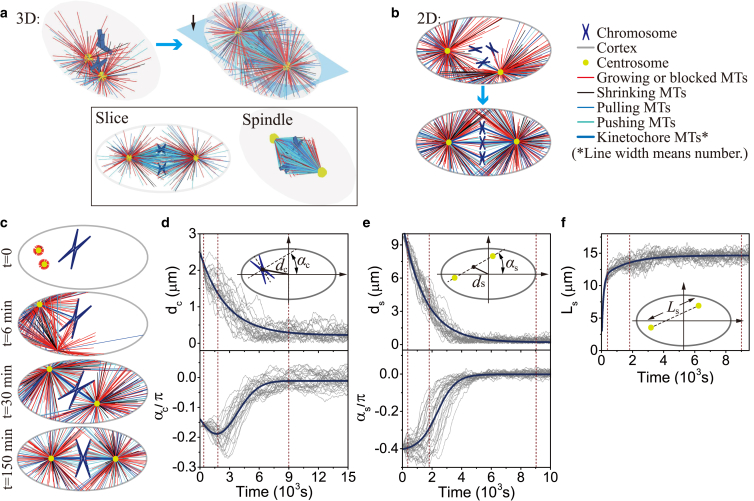
Figure 3.
Self-assembly of mitotic spindles. (a) (Top) Self-assembly of three pairs of chromatids and two centrosomes obtained by 3D simulations (see Fig. S4; Movie S1). (Bottom) The slice view (marked by the black arrow) of the spindle, and the spindle structure with hidden astral MTs. (b) Self-assembly of three pairs of chromatids and two centrosomes obtained by 2D simulations (see Fig. S5; Movie S2). Here, the antiparallel MTs are hidden, and we use the line width to represent the number of kinetochore MTs because they overlap each other. The number of overlapping kinetochore MTs is given in Fig. S7. (c) Self-assembly of one pair of chromatids and two centrosomes obtained by 2D simulations (see Movie S4). The time evolution of (d) chromosome position dc and orientation αc, (e) spindle position ds and orientation αs, and (f) spindle length Ls, is achieved by 20 times of simulations. The parameters used in the simulation are summarized in Table S1. To see this figure in color, go online.