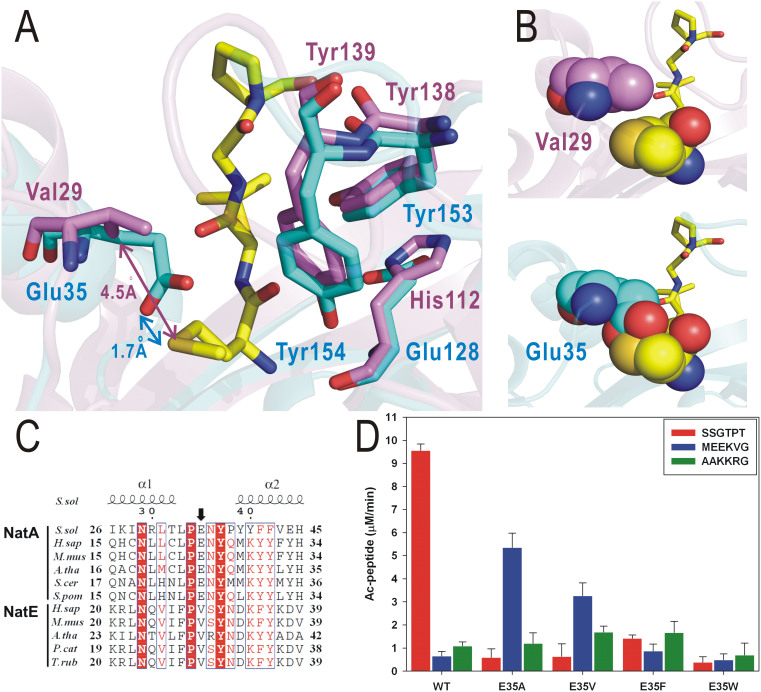
Figure 6. Substrate specificity of SsArd1 (NatA) as compared with hNaa50p (NatE).
(A) Structural superimposition of SsArd1 (cyan) with hNaa50p (magenta) in complex with MLGP (Met-Leu-Gly-Pro) substrate (yellow). (B) Spheres represent the interspace between Val29 of hNaa50p (magenta) and the first residue, Met (yellow) of the substrate from hNaa50p (upper panel) and Glu35 of SsArd1 (cyan) and the first residue, Met (yellow) of the substrate from hNaa50p (lower panel). (C) Sequence alignment of NatA and NatE families from H. sapiens (H.sap), Mus musculis (M.mus), Arabidopsis thaliana (A.tha), Saccharomyces cerevisiae (S.cer), S. pombe (S.pom), Physeter catodon (P.cat) and Takifugu rubripes (T.rub). Sequences were aligned by use of ClustalW2 and the figure was generated by use of ESPript 2.2. Identical and similar residues are labeled by white letters on red backgrounds and red letters, respectively. (D) Enzyme activity assay of wild-type SsArd1 and various single-point Glu35 mutants with different substrate peptides for NatA and NatE.