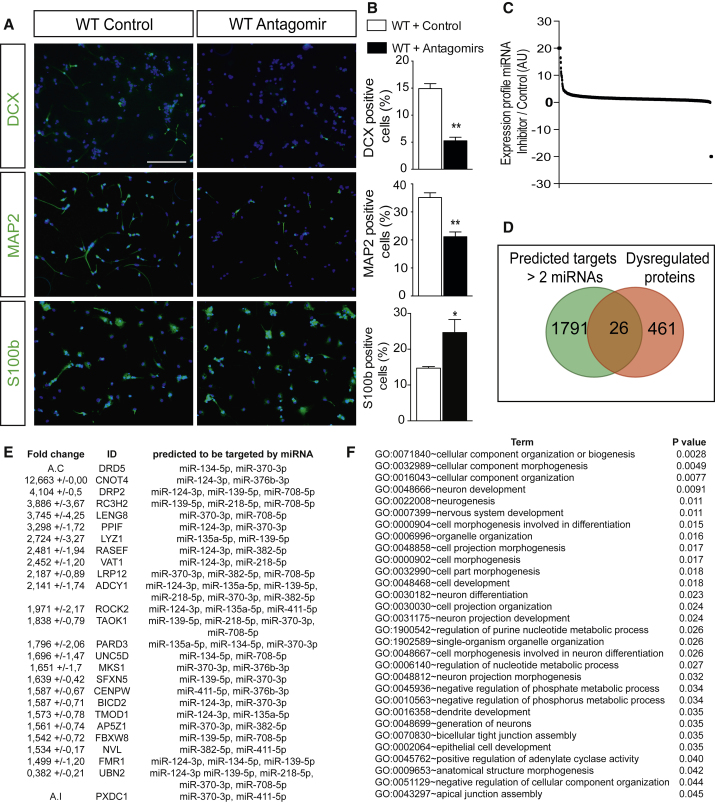
Figure 7.
Convergence of the 11 miRNAs on Neurogenesis-Related Biological Processes
(A) Representative micrographs showing aNSCs from Dicer WT mice transfected with 250 nM scrambled RNA or a pool of miRNA inhibitors (antagomir) (25 nM of each miRNA inhibitor) after 6 DIV of growth factor withdrawal expressing DCX (upper panel), MAP2 (middle panel), and S100b (bottom panel).
(B) Percentage of DCX-, MAP2-, and S100b-positive aNSCs with respect to DAPI in WT aNSCs transfected with scrambled RNA or pool inhibitor.
(C) Expression proteomics profile of miRNA inhibitor with respect to control group at 6 DIV.
(D) Venn diagram showing the putative predicted targets by at least two miRNAs from the in silico analysis that are significantly dysregulated in the proteomics analysis upon miRNA inhibitor administration.
(E) Dysregulated proteins that are putatively targeted by at least two miRNAs. Fold change and SD for WT aNSCs transfected with a pool of inhibitors versus control RNA for each protein expression and predicted targeting miRNAs are shown. A.C, absent in control samples; A.I, absent in miRNA inhibitor samples.
(F) Gene ontology (GO) analysis significantly represented for the 26 predicted common targets dysregulated upon miRNA inhibition.
Data are expressed as mean ± SEM, n = 3 independent experiments containing three replicates. Unpaired t test: ∗p < 0.05, ∗∗p < 0.01. Scale bar, 50 μm.