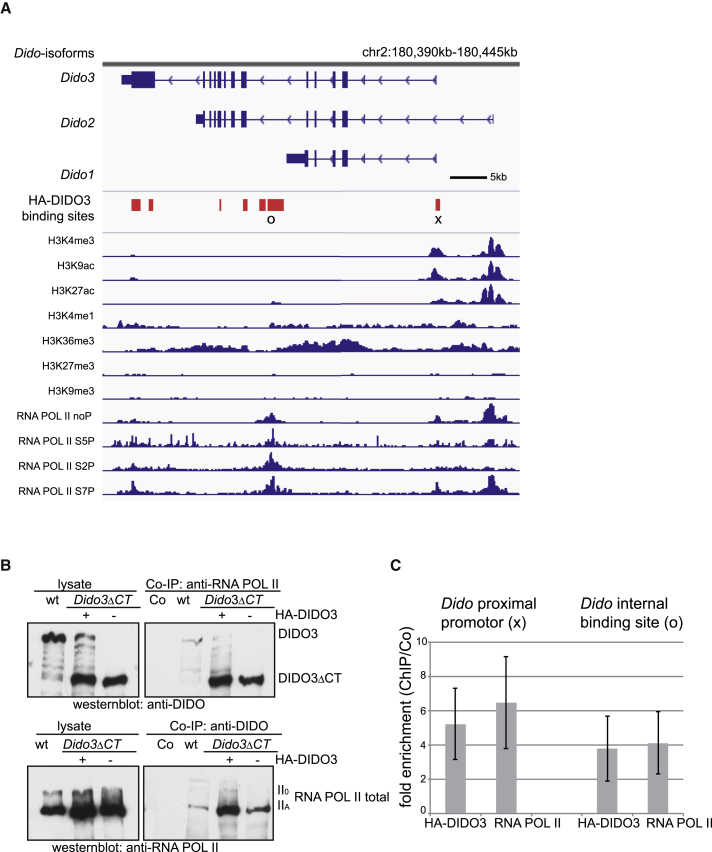
Figure 7.
DIDO3 Binds to the Dido Gene and Interacts with RNA POL II
(A) Image of the 55-kb genomic region containing the three alternatively spliced isoforms of the Dido gene. HA-DIDO3 ChIP-seq binding sites (red boxes); chromatin status is reflected by individual tracks of histone modifications H3K4me3, H3K9ac, H3K27ac, H3K4me1, H3K36me3, H3K27me3, and H3K9me3 (ChIP-seq data from mouse ENCODE project); individual tracks from ChIP-seq data for RNA POL II unphosphorylated, Ser5, Ser2, or Ser7 phosphorylated (Brookes et al., 2012).
(B) Top: DIDO3, HA-DIDO3, and DIDO3ΔCT expression in lysates of WT, mutant ESCs reconstituted with HA-DIDO3, and mutant ESCs (left) and in anti-RNA POL II co-immunoprecipitates (right), developed with anti-DIDO (MAB-1C6). Bottom: expression of total RNA POL II in the same lysates (left) and in anti-DIDO co-immunoprecipitates (right), developed with anti-RNA POL II.
(C) qPCR data demonstrating x-fold ChIP enrichment for anti-HA-DIDO3 and -RNA POL II on Dido proximal promoter (right) and Dido internal sequence after Dido1 3′ UTR (left) relative to negative control (Co; immunoglobulin G), normalized to input amounts. Data are shown as mean ± SD (n from at least three independent experiments).