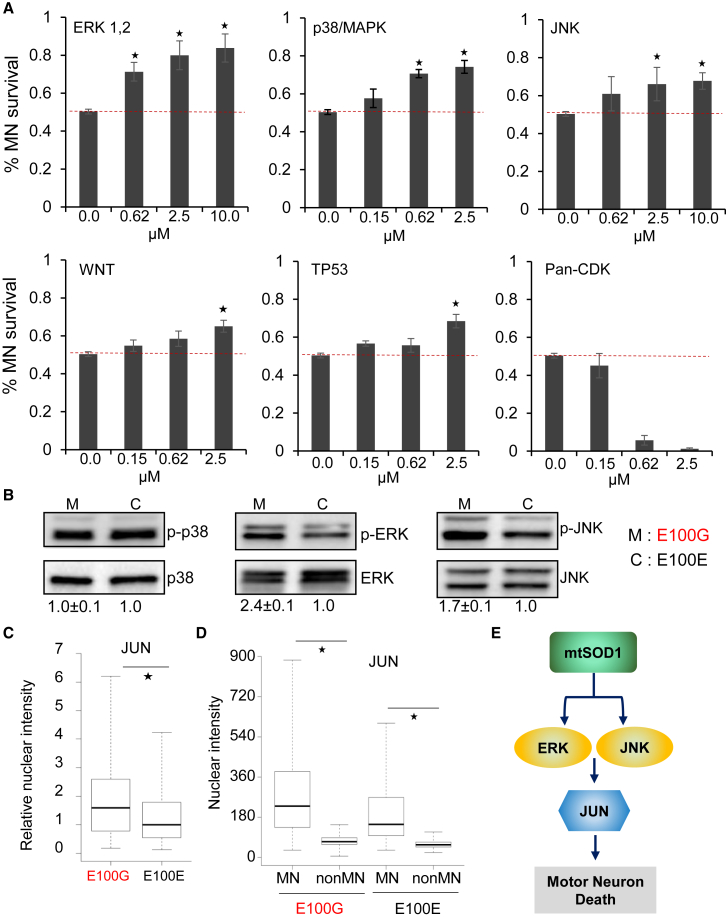
Figure 4.
Identification of Pathways Driving Neurodegeneration in ALS MNs
(A) Quantitation of ISL1-positive mutant SOD1 MNs after treatment with small-molecule inhibitors of the indicated pathways. n = 3, error bars indicate SEM; ∗p < 0.01; p values were estimated using two-tailed Student’s t test.
(B) Western blot analysis showing increased levels of phosphorylated ERK and JNK in mutant SOD1 MNs. Percentages relative to respective isogenic controls are displayed below each blot. Values underlined indicate increased levels of the detected proteins. Lysates from two independent replicates were pooled and assayed in triplicate. Data indicate means ± SEM.
(C) Boxplot displaying nuclear intensities of p-JUN in ALS mutant MNs normalized to the respective isogenic controls. Data from three independent replicates were pooled to generate the boxplot and estimate the p values. ∗p < 0.01; p values were estimated using two-tailed Student’s t test.
(D) Boxplot displaying nuclear intensities of p-JUN in mutant SOD1 MNs and non-MNs as well as isogenic control MNs and non-MNs. Data from three independent replicates were pooled to generate the boxplot and estimate the p values. ∗p < 0.01; p values were estimated using two-tailed Student’s t test.
(E) Schematic depicting pathways activated by mutant SOD1 that drive neurodegeneration.
See also Figure S3.