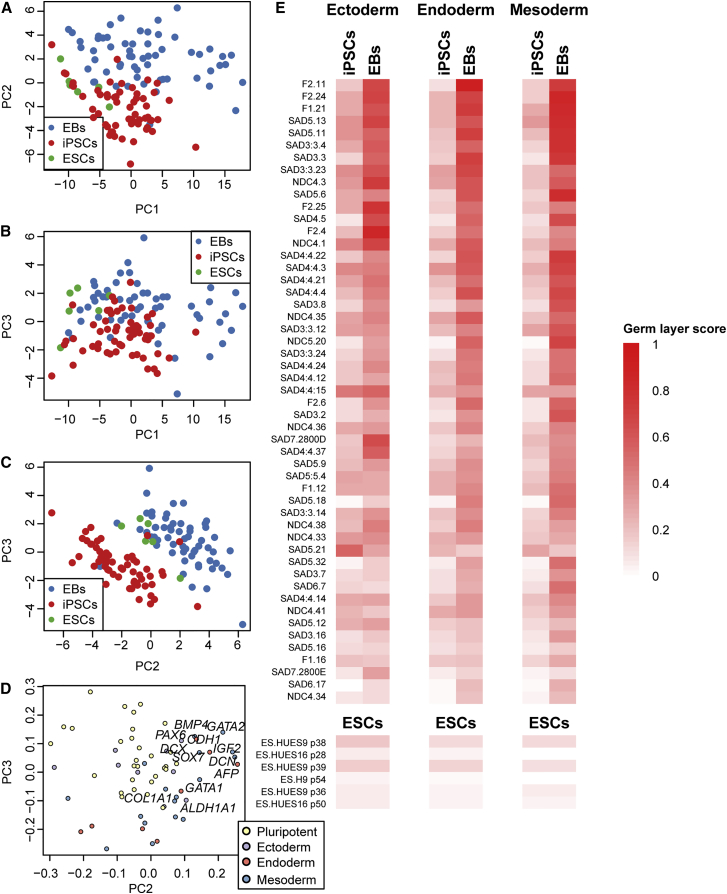
Figure 4.
Germ Layer Scores Show Expression Differences between iPSCs and EBs
(A–C) Principal component analysis of expression of 64 marker genes as measured by qPCR using the Fluidigm Biomark platform: (A) scatterplot PC1-PC2; (B) scatterplot PC1-PC3; (C) scatterplot PC2-PC3. The three plots show that the expression values of the 64 marker genes are able to distinguish iPSCs from embryoid bodies (EBs).
(D) Scatterplot of the weights of each gene on PC2 (x axis) and PC3 (y axis). The 12 genes selected for the qPCR are shown (and are also listed in Table S2). Four genes for each germ layer were chosen because they contributed to the largest expression differences between iPSCs and EBs.
(E) Germ layer scores for iPSCs and EBs (top) and ESCs (bottom) were calculated as the mean value across the four genes in each set. The majority of EBs display high scores for all three germ layers, whereas iPSCs and ESCs have low scores.
See also Table S1. Description of All Lines Used for qPCR, Related to Figure 4, Table S2. List of Primers Used for qPCR, Related to Figure 4, Table S3. Ct Values Detected by qPCR, Related to Figure 4 and Data S1.