Figure 1.
Description of the iPSCORE Cohort
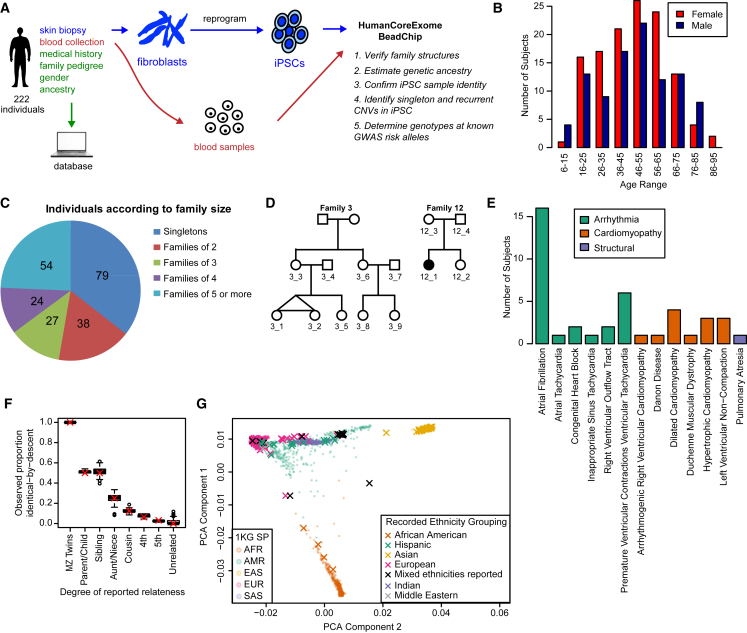
(A) Pipeline for the systematic generation and characterization of 222 iPSC lines. Individuals filled out a questionnaire detailing their medical history, family relationships to other subjects in the cohort, gender, and ancestry. Fibroblasts from skin biopsy were reprogrammed to integration-free iPSC using Sendai virus and frozen at passage 12. Genomic DNA isolated from the iPSC and the subject-matched blood samples were hybridized to the HumanCoreExome array. The resulting data were then used to confirm reported family structure, reported ancestry, and iPSC sample identity (match with blood sample), and to perform CNV analysis (iPSC characterization) and determine status of known disease risk alleles.
(B) Age distributions of males and females.
(C) Pie chart showing how many individuals are singletons or in a family size of 2, 3, 4, and 5 or more.
(D) Pedigrees of two representative families; numbered individuals indicate presence in the study. Family 3 is a two-generation family with identical twins (nine subjects), and Family 12 has a member diagnosed with ventricular tachycardia and congenital heart block (four subjects).
(E) Number of individuals with cardiac disease, grouped by disease type. Some individuals are affected by multiple types of arrhythmia.
(F) Boxplot showing the observed proportion of the genome identical by descent (pIBD) as a function of the reported family relationship. The box hinges indicate the 25th and 75th quantiles and the whiskers extend to 1.5 times the interquartile range. A red “X” indicates the expected mean pIBD given the number of generations that separate the individuals.
(G) An x-y plot showing the first versus second components of a principal component analysis using genotype data from a subset of SNPs present on the array mapped onto a principal component analysis from the 1,000 Genomes Project (1KG) super populations (SP) (small faded circles). Individuals from the iPSCORE cohort are mapped onto these components with their recorded ethnicity grouping shown by a colored X.