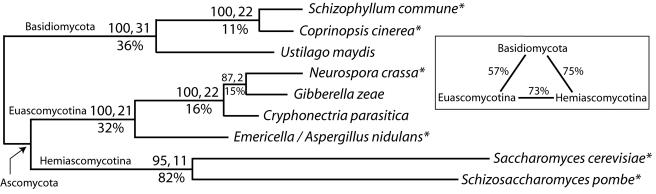
FIG. 2.
Phylogenetic analysis under the parsimony criterion of RGS protein sequences from nine taxa within Ascomycota and Basidiomycota. Taxa are referred to by their Linnaean names (see Materials and Methods for synonyms and gene names). Those genera followed by asterisks were also sampled in Bruns et al. (4). Number of most-parsimonious trees, 1; most-parsimonious tree length, 1006; number of parsimony-informative characters, 221; total number of included characters, 389; consistency index, 0.9324; retention index, 0.8265. Bootstrap percentages followed by Bremer support/decay indices are shown above the internal branches. The percent uncorrected average pairwise amino acid differences are shown below branches and in the enclosed box. Branch lengths are proportional to the number of amino acid changes and are optimized on a topology rooted between Ascomycota and Basidiomycota.