Abstract
Oxytocin appears beneficial for autism spectrum disorder (ASD), and more than 20 single-nucleotide polymorphisms (SNPs) in oxytocin receptor (OXTR) are relevant to ASD. However, neither biological functions of OXTR SNPs in ASD nor critical OXTR SNPs that determine oxytocin’s effects on ASD remains known. Here, using a machine-learning algorithm that was designed to evaluate collective effects of multiple SNPs and automatically identify most informative SNPs, we examined relationships between 27 representative OXTR SNPs and six types of behavioral/neural response to oxytocin in ASD individuals. The oxytocin effects were extracted from our previous placebo-controlled within-participant clinical trial administering single-dose intranasal oxytocin to 38 high-functioning adult Japanese ASD males. Consequently, we identified six different SNP sets that could accurately predict the six different oxytocin efficacies, and confirmed the robustness of these SNP selections against variations of the datasets and analysis parameters. Moreover, major alleles of several prominent OXTR SNPs—including rs53576 and rs2254298—were found to have dissociable effects on the oxytocin efficacies. These findings suggest biological functions of the OXTR SNP variants on autistic oxytocin responses, and implied that clinical oxytocin efficacy may be genetically predicted before its actual administration, which would contribute to establishment of future precision medicines for ASD.
Keywords: imaging genetics, neuropeptide, pervasive developmental disorder, pharmacogenetics, randomized clinical trial, support vector machine
Introduction
Autism spectrum disorder (ASD), a prevalent neurodevelopmental disorder characterized by impaired socio-communicational interactions and repetitive restricted behaviors, have polygenetic backgrounds ranging from heritable genetic variations to de novo mutations and copy number variants (Devlin and Scherer, 2012; Meyer-Lindenberg and Tost, 2012; Lai et al. (2014)). Variations in the oxytocin receptor gene (OXTR) are among such key genetic factors (Meyer-Lindenberg and Tost, 2012; Yamasue, 2013; LoParo and Waldman, 2015), and previous researches reported associations between ASD and more than 20 single-nucleotide polymorphisms (SNPs) in OXTR (Wu et al., 2005; Ylisaukko-oja et al., 2006; Jacob et al., 2007; Lerer et al., 2008; Yrigollen et al., 2008; Liu et al., 2010; Wermter et al., 2010; Yamasue, 2013; LoParo and Waldman, 2015).
This ASD-OXTR relationship is supported by empirical evidence for seemingly beneficial effects of oxytocin on this developmental disorder. Although further confirmatory studies with a large-scale sample are necessary (Shen, 2015), clinical trials reported that single-dose intranasal oxytocin administration mitigated autistic social behaviors and neural responses to socio-communicative information (Hollander et al., 2007; Guastella et al., 2010; Aoki et al., 2014; Domes et al., 2014; Watanabe et al., 2014a; Auyeung et al., 2015). Moreover, behavioral and neuroimaging studies employing neurotypical individuals added indirect evidence through identification of behavioral/neural links between several OXTR SNPs and ASD-associated social tendencies (Rodrigues et al., 2009; Inoue et al., 2010; Tost et al., 2010; Furman et al., 2011; Yamasue et al., 2011; Guastella et al., 2012; Kumsta and Heinrichs, 2013). Collectively, these observations suggest that some OXTR variants in ASD individuals would alter their oxytocin-related neural circuits and partly induce their socio-communicational dysfunctions (Meyer-Lindenberg and Tost, 2012; Yamasue, 2013; LoParo and Waldman, 2015)
However, which OXTR SNPs crucially induce such biological effects on neural mechanisms underlying atypical social cognitions in ASD is still unclear (Brüne, 2012), and thus it remains unknown which OXTR SNPs significantly regulate oxytocin’s effects on the social symptoms of the disorder. Although a recent study examined associations between three OXTR SNPs and oxytocin response in neurotypical adults (Montag et al., 2013), no previous study employing ASD individuals has examined such relationships between oxytocin efficacy and genetic variability. In addition to such a biological significance, given a large variety of autistic responses to continual oxytocin administration (Anagnostou et al., 2012; Dadds et al., 2014; Guastella et al., 2015; Watanabe et al., 2015; Yatawara et al., 2016), it is also clinically essential to identify genetic variants of OXTR that enhance/deteriorate oxytocin’s beneficial effects on ASD (Kumsta and Heinrichs, 2013; Yamasue, 2013).
Here, we attempted to determine such biologically and clinically crucial OXTR SNPs that significantly affect oxytocin efficacy in ASD by applying a machine-learning algorithm to original datasets consisting of individual genetic information and behavioral/neural responses to the neuropeptide. The dataset about the oxytocin efficacy was extracted from our previous reports about a randomized, double-blind, placebo-controlled, cross-over within-participant trial (Watanabe et al., 2014a; Aoki et al., 2015). We employed not a univariate but multivariate analysis because it is crucial to evaluate collective effects of multiple SNPs in investigation of multigene disorders including ASD (Li et al., 2012; Miao-Xin Li, 2011; Wu et al., 2011). The machine-learning-based approach (here, support vector regression, SVR) was selected because, compared with a conventional general linear model, the technique can yield results that are more robust against noise and outliers, and have larger generalization capability (Drucker et al., 1997; Thissen et al., 2004). Moreover, like deep learning methods (Hinton and Salakhutdinov, 2006), the current algorithm was designed to automatically select the most informative SNP sets without specific a priori assumptions.
Using these data and algorithm, we examined a hypothesis whether the behavioral and neural efficacy of oxytocin was predicted by patterns of specific OXTR SNP sets. Consequently, we identified the most informative combinations of OXTR SNPs for different types of oxytocin efficacy, and confirmed the different SNP sets could predict different oxytocin responses. The results implied distinct biological functions between several prominent OXTR SNPs.
Materials and methods
Overall analysis design
The current analysis procedure mainly consisted of the following six steps. (i) First, we excluded OXTR SNPs whose patterns were inappropriate for regression analysis (Figure 1A). (ii) Second, we prepared six indices of behavioral and neural oxytocin effects that had been already measured in our previous clinical trial (Watanabe et al., 2014a; Aoki et al., 2015). (iii) Third, using the machine-learning-based regression analysis (i.e., SVR), we calculated the associations between the oxytocin efficacy and all the possible sets of the OXTR SNPs, and searched for the most informative SNP sets (Figure 1C). (iv) Afterwards, we tested the robustness of the SNP selections by changing several analysis parameters. (v) Fifth, based on the selected informative SNP sets, we directly examined how accurately the SNP sets could predict the oxytocin efficacy. (vi) Finally, we evaluated the weight values assigned to the selected SNPs in the regression analysis, and examined whether the SNPs enhanced or deteriorated beneficial effects of oxytocin in high-functioning Japanese adult ASD males.
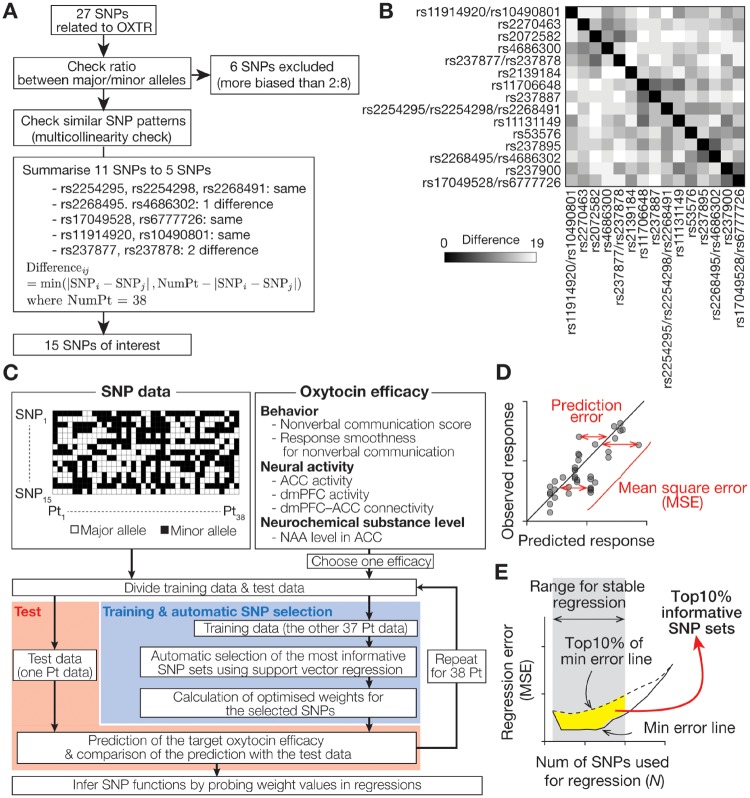
Fig. 1.
(A) Selection of SNPs of interest. Based on an established database (Affymetrix Genome-Wide Human SNP Array 6.0) and previous studies (Yamasue, 2013; LoParo and Waldman, 2015), 27 OXTR SNPs were identified for 38 participants with ASD (Supplementary Table 1). First, six SNPs were excluded because of their unbalanced ratios between their major and minor alleles across participants (more biased than 2:8), which could cause inaccurate machine-learning-based analysis. This 2:8 threshold is more conservative than that in previous literature (Johnson et al., 2009; Watanabe et al., 2011). Second, we examined multicollinearity between the remaining 21 SNPs, and 11 SNPs were summarized to five patterns. The patterns of rs2268495 and rs4686302 were summarized to that of rs2268495 because the rs2268495 pattern was less biased than the rs4686302 pattern. Using this logic, the patterns of rs237877 and rs237878 were summarized to that of rs237878. Finally, 15 OXTR SNPs were selected (Table 1). (B) Difference in across-participant SNP patterns. The remaining 15 OXTR SNPs showed ≥ 7 differences from each other, which is supposed to assure sufficient independency of these SNP patterns for the following regression analysis. (C–E) Analysis procedure. Using the 15 OXTR SNPs, we performed machine-learning-based regression analyses to determine specific SNP sets with sufficient information to predict behavioral and neural oxytocin efficacy, which had been measured in our previous trials (Watanabe et al., 2014a; Aoki et al., 2015). Six types of oxytocin efficacy was defined as (i) non-verbal communication score = increased number of NVJ (oxytocin–placebo); (ii) response smoothness = decreased reaction time for NVJ (placebo–oxytocin); (iii, iv) ACC/dmPFC activity = increased brain activity for NVJ in ACC/dmPFC (oxytocin–placebo; ACC, anterior cingulate cortex; dmPFC, dorsomedial prefrontal cortex); (v) dmPFC–ACC connectivity = increased NVJ-specific functional connectivity from dmPFC to ACC (oxytocin–placebo); (vi) NAA level in ACC = increased level of NAA in ACC (oxytocin–placebo).; NAA, N-acetylaspartate. For each of the six types of oxytocin response, we conducted machine-learning-based automatic selections of the most informative SNP sets, and examined its predictability using an independent test dataset. Briefly, we ranked SNPs based on MSEs of the regressions using them (panel D), and extracted the most informative SNPs based on the probability of each SNP appearing in top 10% least MSE SNP groups (panel E; see Supplementary Figure S1 for details).
Participants
The current participants were the same as those in our previous clinical trials (Watanabe et al., 2014a) (i.e. ethnically homogeneous 38 high-functioning ASD males with age ≥ 20 and FIQ > 80, 35 of which were non-medicated). The original ample size of the trial (n = 40) was determined by a power analysis based on our previous study about the relationship between an OXTR SNP (rs2254298) and the amygdalar volume in ethically the same cohort as the current study (Inoue et al., 2010; see Supplementary Materials). Two of the 40 original participants were excluded because their behavioral responses were not recorded due to technical problems in the trial.
Their ASD diagnoses were made by experienced psychiatrists (HY/HK) with the strict criteria of DSM-R IV after more than 2 months of follow-up examinations, and were validated by certified psychiatric psychologists with Japanese version of ADI-R (HK) (Lord et al., 1994) and/or ADOS (Dr Miho Kuroda) (Lord et al., 1989). All the participants had sufficiently high IQs ranging from average to above average in the full scale of WAIS-R, Japanese version. The study protocol was registered in University Hospital Medical Information Network clinical trial registry (UMIN000002241/000004393). Written informed consent was obtained from all the participants.
OXTR genotyping
Among 27 OXTR SNPs of interest, 21 SNPs were selected based on Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA; non-* SNPs in Supplementary Table S1), whereas the other six SNPs were included according to previous reports (Yamasue, 2013; LoParo and Waldman, 2015; * SNPs in Supplementary Table S1). TaqMan genotyping platform was used to genotype these SNPs (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA; see Supplementary Materials).
Step 1: exclusion of SNPs inappropriate for regression analysis
First, we excluded OXTR SNPs whose distribution patterns across participants could induce inaccurate calculations in the following analyses (Figure 1A). Technically, SNPs whose across-participant distributions were too biased (here, a major allele is seen in >80% of the sample) were removed because they could cause overfitting in machine-learning procedures (Bishop, 2006). This threshold is more conservative than those adopted in previous machine-learning-based studies (Johnson et al., 2009; Watanabe et al., 2011).
Second, we examined multicollinearity (Kumar, 1975) between the remaining SNPs by calculating the difference in the across-participants SNP patterns, which was defined as min(|SNPi – SNPj|, Nparticipant – |SNPi – SNPj|), where Nparticipant denotes the number of participants. SNPs whose differences were ≤ 2 were summarized into one of them (Figure 1B). Consequently, allelic information of 15 OXTR SNPs was used in the following analysis (Table 1).
Table 1.
SNPs of interest
| SNP | Allele assigned to 2 | Allele assigned to 1 |
|---|---|---|
| rs11914920/rs10490801 | AA/TT | G carrier/C carrier |
| rs2270463 | GG | A carrier |
| rs2072582 | GG | A carrier |
| rs4686302 | CC | T carrier |
| rs237877/rs237878 | CC/TT | T carrier/C carrier |
| rs2139184 | CC | T carrier |
| rs11706648 | AA | G carrier |
| rs237887 | GG | A carrier |
| rs2254295/rs2254298/rs2268491 | TT/GG/CC | C carrier/A carrier/T carrier |
| rs11131149 | GG | A carrier |
| rs53576 | GG | A carrier |
| rs237895 | TT | C carrier |
| rs2268495/rs4686302 | GG/CC | A carrier/T carrier |
| rs237900 | GG | A carrier |
| rs17049528/rs6777726 | GG/GG | A carrier/A carrier |
Step 2: identification of six oxytocin efficacy
The data for the six types of oxytocin efficacy (Figure 1C) were extracted from our previous studies (Watanabe et al., 2014a; Aoki et al., 2015).
Five of the six responses were measured using a social cognition task that was designed to recruit cognitive ability to smoothly resolve conflicts between verbal and non-verbal social information (Watanabe et al., 2012; 2014b). This ability is implicitly used to understand jokes and satires in typically developing (TD) individuals.
In this task, participants were presented with a series of short videos. In each video, a professional actor spoke an emotional phrase (positive/negative verbal information) with emotional facial and vocal expression (positive/negative non-verbal information), and the participants were asked to judge whether the actor in the video felt like a friend or foe.
By focusing on responses to movie stimuli whose verbal information was incongruent with non-verbal information (e.g. a positive phrase was spoken with negative facial and voice expression), we first characterized individual social behaviors. For example, a ‘friend’ judgment for an actor speaking a negative phrase with positive facial and vocal expressions was regarded to be a decision mainly based on non-verbal information, whereas a ‘foe’ judgment for the same actor was regarded as verbal-information-based judgments. Afterwards, we estimated brain activity specific to such non-verbal-information-based judgments (NVJs), and identified brain mechanisms underlying atypical social behaviors in ASD individuals.
In fact, this social cognitive task enabled us to detect ASD-specific behavioral and neural responses that were linked with the severity of the social symptoms of autism (Watanabe et al., 2012). When compared with TD individuals, ASD individuals were more likely to emphasize verbal information during such friend/foe judgments. The number of NVJs was significantly less in ASD group than in TD group, and the reaction time for NVJs was also longer in ASD individuals. In addition, during this NVJ behavior, ASD individuals showed significantly less brain activity in two medial prefrontal regions than TD individuals (anterior cingulate cortex, ACC; dorsomedial prefrontal cortex, dmPFC). Notably, these behavioral and neural responses were correlated with the severity of the socio-communicational symptoms of ASD.
Moreover, this task allowed us to detect five behavioral and neural effects of single-dose intranasal oxytocin (24IU) on social symptoms of autism (Watanabe et al., 2014b). When compared with placebo, single-dose oxytocin significantly increased the number of NVJs and reduced the reaction time for NVJs with recovering brain activity of the two brain regions (ACC and dmPFC) and bidirectional functional connectivity between the two regions. Furthermore, using this task, we have recently reported significant behavioral and neural effects of oxytocin even in a long-term continual administration (Watanabe et al., 2015).
Note that in our previous clinical trial (Watanabe et al., 2014b), we had attempted to reduce behavioral and neural effects of practice in this type of stroop task by (i) introducing sufficient training sessions before the MRI scanning and (ii) randomizing the order of drug administration. Regarding (i), the participants were asked to undergo more than 20 ‘friend or foe’ judgment trials with movie stimuli that were not included in the following MRI experiments. This training session were expected to saturate behavioral/neural responses to the tasks in each participant, and hence reduce the practice effects within day and within participant. Regarding (ii), practice effects over the different experiment days were supposed to be controlled by randomizing drug administration days across participants (i.e. randomization of oxytocin and placebo days in a crossover design).
Given these previous observations, it is reasonable to assume that these five types of behavioral and neural response are sensitive and specific enough to represent oxytocin’s efficacy on autistic social symptoms. To reduce confounding effects of selection bias, we used all of them as outcome measures to investigate relationships between types of OXTR SNP and oxytocin efficacy as follows (Figure 1C): (i) non-verbal communication score = increased number of NVJs (oxytocin–placebo); (ii) response smoothness = decreased reaction time for NVJ (placebo–oxytocin); (iii, iv) ACC/dmPFC activity = increased brain activity for NVJs in ACC/dmPFC (oxytocin–placebo); (v) dmPFC–ACC connectivity = increased NVJ-specific functional connectivity from dmPFC to ACC (oxytocin–placebo).
In addition, because oxytocin affected the neurochemical substance level (Aoki et al., 2015), (vi) increased level of N-acetylaspartate (NAA) in ACC (oxytocin–placebo) was adopted as the sixth oxytocin efficacy. To reduce effects of outliers on the following regressions, these oxytocin efficacy values were normalized.
Step 3: search for informative SNP sets with SVR
We then searched for the informative SNP sets that accurately predicted each oxytocin efficacy by implementing a SNP ranking algorithm based on a ν-linear SVR (Bishop, 2006; Long et al., 2011) (Figure 1C, Supplementary Figure S1) in MATLAB R2014b (MathWorks) with LIBSVM package (www.csie.ntu.edu.tw/∼cjlin/libsvm/). We used a linear SVR rather than non-linear one to avoid overfitting and enable straight-forward interpretation of weight values later (Bishop, 2006).
First, the SNP information was binarized (major/minor allele as 2/1; Table 1). After choosing one of the six oxytocin efficacy indices, we divided the SNP and response data to test data (consisting of one participant data) and training data (consisting of the others’ data) in a leave-one-out-cross-validation (LOOCV) manner (Bishop, 2006).
Second, using the training data, we fitted the binary information of certain n SNPs (1 ≤ n ≤ 15) to the oxytocin efficacy using a SVR, and calculated the mean square error (MSE) of the regression (Figure 1D; Long et al., 2011). In each SNP number (n), we repeated this fitting and MSE calculation for all the possible 15!/n!(15–n)! SNP sets, and ranked the SNP sets according to the MSE ascending order (‘SNP Ranking’ in Supplementary Figure S1).
We did not directly adopt the SNP set with the smallest MSE as the most informative SNP combination, because the MSE calculations can be noisy owing to the current limited dataset size and may be simply a result of overfitting (Bishop, 2006). Instead, we first calculated the probability of how often each SNP appeared in the Top10% most informative SNP sets (i.e. Top10% of rankings; Figure 1E). Moreover, to reduce effects of inaccurate calculations due to so-called ‘curse of dimension’ (i.e. here, imprecise estimations at a large n; Bishop, 2006), we focused on a range whose MSE calculations were relatively stable (i.e. 1 ≤ n ≤ 10; Figures 1E and 2A), and averaged the appearance probability within the range. We finally selected sets of the most informative SNPs based on this average appearance probability (‘SNP Selection’ in Supplementary Figure S1). These selection procedures were anticipated to improve the robustness of the SNP selection.
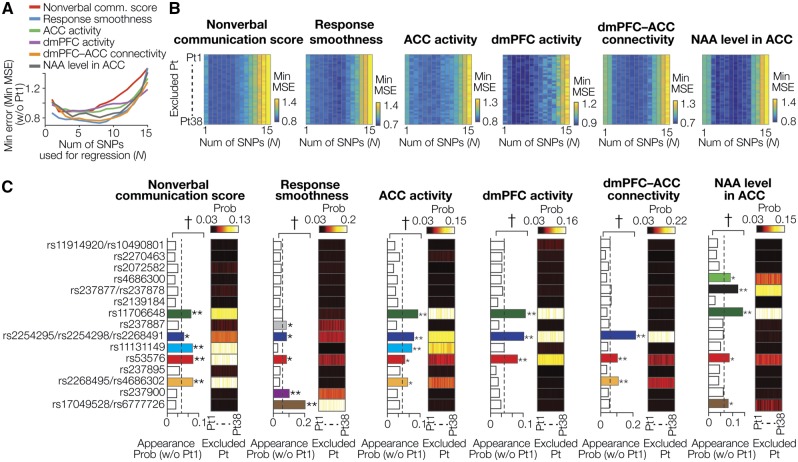
Fig. 2.
Informative SNPs. (A, B) Minimum error and size of SNP sets. Panel A shows representative relationships between the number of SNPs used for the SVR (n) and the minimum (min) MSE, which were estimated using data excluding those obtained from Participant no. 1 (Pt1). Panel B shows associations between the SNP number and min MSE in all patterns of training data sets. The vertical axes show ID of the participants whose data were excluded from the training data. For all the six types of oxytocin efficacy, the min MSE was relatively stable between n = 1 and 10, whereas in n > 10, the min MSE gradually increased. MSE, mean square error. (C) Probability of SNP appearance in the Top10% informative SNP sets. The bar graphs show how often each SNP was included in the Top10% informative SNP groups when the analysis used data excluding those of Pt1. The neighboring color boxes show the appearance probability patterns in all the training data sets. The appearance probabilities were calculated as an average across 1 ≤ n ≤ 10. * and ** represent Bonferroni-corrected P values (* <0.05; **< 0.01) in residual tests following chi-squared tests. †shows that the distribution of the appearance probability is significantly rare in post-hoc permutation tests (PBonferroni-corrected < 0.05). The dash lines represent the value of by-chance appearance probability (i.e. 1/15 = 0.067). Colored bars indicate the significantly informative SNPs. For every type of the oxytocin efficacy, the informative SNP sets with significantly high appearance probability were the same across all the 38 different training datasets
Distributions of the appearance probability were evaluated by chi-squared tests and post-hoc residual tests, which was further validated by permutation tests with 10 000 random permutations.
Statistical evaluations were corrected in Bonferroni manner (for the chi-squared tests across six oxytocin effects, α= 0.05/6; for the post-hoc residual test across 15 SNPs, α= 0.05/15; for the permutation tests across six oxytocin effects, α= 0.05/6). This Bonferroni correction may be too conservative because the five oxytocin efficacy indices were likely to be correlated with each other (absolute value of Fisher-transformed correlation coefficients: ∣Z∣ = 0.47 ± 0.22, mean ± SD). However, given the current relatively limited sample size, we attempted to reduce the risk of overestimation by adopting this conservative correction method.
Step 4: tests of the robustness of the informative SNPs selections
We then examined the robustness of the SNP selections by changing (i) the definition of the top informative SNP sets and (i) the SNP number (n) used to calculate the average appearance probability. To test (i), we repeated the SNP selection procedure by changing the definition of the top informative SNP groups from Top5% to Top20% by one point (Supplementary Figure S3A). To test (ii), we repeated the same analysis by changing the range of n from 1 ≤ n ≤ 10 to 5 ≤ n ≤ 10 (Supplementary Figure 4A) and moving the range from 1 ≤ n ≤ 5 to 6 ≤ n ≤ 10 (Supplementary Figure S5A).
Step 5: direct evaluation of predictability of the selected SNPs
We then directly evaluated how accurately the selected SNP sets predicted the oxytocin efficacy. The predictability was estimated in a LOOCV manner. For each oxytocin efficacy, we determined regression weights of the informative SNPs using training data (datasets recorded from 37 individuals), applied the weights to the test SNP data (a dataset derived from the remaining one individual), and calculated the predicted value of the oxytocin efficacy. We repeated these procedures for every training dataset (i.e. 38 different datasets), and obtained efficacy prediction for all the 38 individuals. We then evaluated the similarity between these predicted and real responses by performing F tests on the fitness of the regressions and by calculating Pearson correlation coefficients between them.
We also examined the accuracy in distinguishing individuals with high oxytocin sensitivity from those with relatively less sensitivity by counting the number of the participants for whom predicted responses had the same directions as empirically observed responses (i.e. both > 0 or both < 0; participants in the light gray areas in Figure 3B). Such classification using ‘zero’ as a threshold was justified because the data were normalized prior to the SVR calculations.
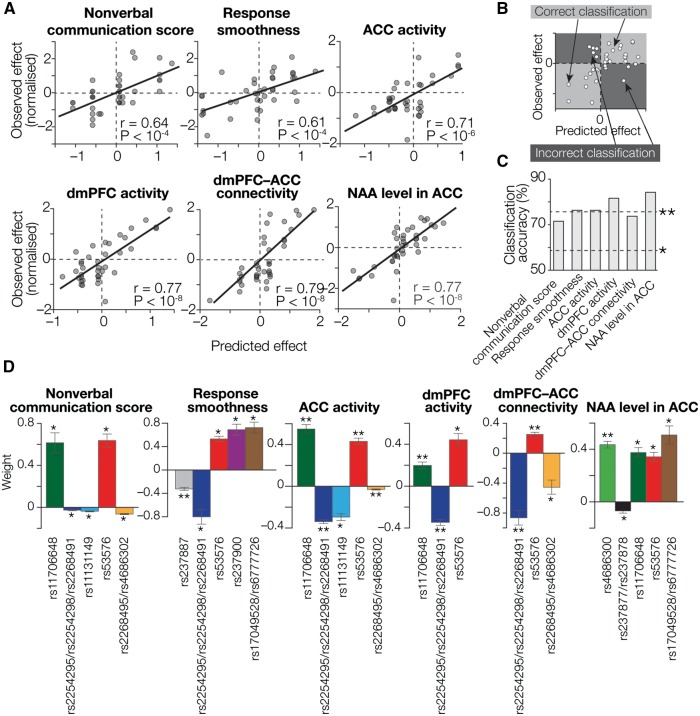
Fig. 3.
Prediction of oxytocin efficacy based on OXTR SNPs. (A) Regression accuracy. The X axes show predicted oxytocin efficacy calculated by SVR using the most informative SNP sets (colored SNPs in Figure 2C), whereas the Y axes show normalized values of the empirically observed effects. Each circle represents a participant. For all the oxytocin effects, we observed significantly high correlations between the predicted and empirical values (PBonferroni-corrected < 0.05; Supplementary Table S2). To reduce the effect of outliers, all the indices of oxytocin efficacy were normalized prior to the regression analysis. (B, C) Based on these regression analyses, we calculated accuracy of this machine-learning technique for classifying participants into an oxytocin-responsive group and a relatively irresponsive group. As schematically shown in panel B, participants plotted in the light gray areas are regarded as correctly classified ones, whereas those in the dark gray areas are not. For all six types of oxytocin efficacy, the classification accuracy was significantly higher (*PBonferroni-corrected < 0.05; **PBonferroni-corrected < 0.01 in binominal tests; panel C). (D) Weight values assigned to the informative SNPs. We examined the weight values assigned to the informative SNPs by the SVR analyses. The Y axes represent weight values averaged across four independent SVR calculations using four independent datasets. Notably, all the informative SNPs show consistent directions across different types of oxytocin efficacy (e.g. weights assigned to rs53576 are significantly positive in all the responses). *PBonferroni-corrected < 0.05; **PBonferroni-corrected < 0.01, in one-sample t-tests. Error bars, SEM. See also Table 2.
These statistical evaluations were corrected in Bonferroni method (α= 0.05/6).
Step 6: examination of functionality of the selected SNPs
Finally, we examined weights assigned in the linear SVR. To ensure the independence between the estimated weights, we first randomly divided the 38 participants into four groups (n = 9, 9, 10 and 10). Within each group, we performed the same SNP selections for the six oxytocin efficacy. After confirming that the appearance probability of the originally selected informative SNPs was above a chance level in every group, we estimated regression weights for the selected SNPs. Using these independently calculated four weights for each informative SNP, we conducted one-sample t-tests and examined whether each weight was significantly different from zero.
Multiple comparisons in these tests were corrected in Bonferroni method (α= 0.05/[number of the informative SNPs for each type of oxytocin efficacy]).
Results
Exclusion of SNPs inappropriate for regression analysis
We first excluded six of the 27 OXTR SNPs whose major allele types were too dominant across participants (>80% of the sample) and could cause inaccurate regression analysis (Figure 1A, Supplementary Table S1). Then, we summarized 11 of the remaining 21 SNPs into five patterns because of their high multicollinearity, and obtained 15 SNPs (Table 1; see also legends for Figure 1A), whose distributions were relatively unbiased and sufficiently different from each other (difference ≥ 7, variance inflation factor < 1.9; Figure 1B). This procedure to avoid high multicolinearity is qualitatively equivalent to a protocol to assure linkage disequilibrium between the SNPs (Supplementary Figure S2). In the following analysis, we investigated associations between the oxytocin efficacy and patterns of these 15 OXTR SNPs.
Selection of the most informative SNP sets
Next, using the 15 SNPs, we searched for the most informative SNP combinations that could robustly and accurately predict individual oxytocin efficacies. For this purpose, we evaluated the probability of each SNP appearing in Top10% informative SNP sets (Figure 1C, Supplementary Figure S1). Because the regression errors (i.e. minimum MSEs) were relatively stable when using 1–10 SNPs for all the oxytocin effects in all the patterns of the training datasets (Figure 2A and B), we calculated the appearance probability based on these stable results of the regressions analysis.
As a result, we first confirmed that the distributions of the appearance probability across 15 SNPs were significantly skewed compared with a uniform distribution for every oxytocin efficacy (x2(14) > 118, PBonferroni-corrected < 0.001 in chi-squared tests; Figure 2C). Then, we found that the appearance probabilities of several specific SNPs were significantly higher than the chance level (SNPs indicated with colored bars in Figure 2C;Z > 4.1, PBonferroni-corrected < 0.05 in posthoc residual tests; P < 0.0075, PBonferroni-corrected < 0.05 in posthoc permutation tests). These results suggest that such SNPs with significant high appearance probability can be regarded as the most informative SNP sets.
To avoid circular analysis, these entire SNP selection procedures were conducted after dividing the dataset to training and test datasets according to a LOOCV manner (Figure 1C, Supplementary Figure S1). Notably and coincidently, the informative SNP sets showing significantly high appearance probability were the same across all the 38 different training datasets for every oxytocin response (color boxes in Figure 2C).
Examination of robustness of SNP selections
We then examined the robustness of these SNP selections by changing several analysis parameters. First, we repeated the SNP selection procedure with changing the definition of ‘top informative groups’ from Top5% to Top20% lowest MSE groups by one point (Supplementary Figure S3A). Consequently, for every oxytocin efficacy, all of the most informative SNPs originally selected in Figure 2C were consistently identified as being significantly informative (PBonferroni-corrected < 0.05 in residual tests) in >85% of the parameter changes, whereas the other SNPs were not selected so often (≤ 50% of the changes; Supplementary Figure S3B and C). These results show the robustness of the original SNP selections against changes of the definition of ‘top informative groups’.
Second, we also confirmed the robustness of the SNP selections by changing the SNP number range (n) used for calculations of the appearance probability (Supplementary Figure S4A). Even after the range width was changed from 10 to five, all the original informative SNPs shown in Figure 2C were always selected as informative ones, whereas the other SNPs were not so consistently chosen (≤40% of the changes; Supplementary Figure S4B and C). Moreover, this robustness was still observed when we moved the window for the target SNP number form 1 ≤ n ≤ 5 to 6 < n ≤ 10 (Supplementary Figure S5A): for every oxytocin efficacy, the originally-selected SNPs were almost always significantly informative (>83% of the changes), whilst the other SNPs were not so consistently selected (<32% of the changes; Supplementary Figure S5B and C). These results suggest the robustness of the original SNP selections against modification of the manners to calculate ‘appearance probability’.
Direct estimation of prediction accuracy
Next, using these robustly selected informative SNP selections, we directly examined how accurately these SNP sets could predict oxytocin efficacy. We first found that the regressions using the selected SNP sets were well fitted to every type of oxytocin response (PBonferroni-corrected < 0.05 in F tests, adjusted R2 > 0.27; Supplementary Table S2), and confirmed that the effects predicted in the regressions were significantly correlated with the empirically observed effects (r ≥ 0.61, PBonferroni-corrected < 0.05; Figure 3A). Note that the independence between the training and test data was preserved here, because the same SNP sets were selected as the most informative SNP sets across all different training datasets for each oxytocin response (Figure 2C).
Moreover, these informative SNP sets enabled us to distinguish individuals who were above-average responsive to oxytocin from those who were below-average responsive to the neuropeptide (>71% accuracy, PBonferroni-corrected < 0.05 in binominal tests; Figures 3B and C, Supplementary Table S3). Furthermore, such accurate predictions of the oxytocin efficacies were consistently achieved regardless of the order of the administration of oxytocin and placebo (r ≥ 0.57; Supplementary Figure S6).
Taken together, these results show that the selected OXTR SNP sets can accurately predict behavioral and neural oxytocin responses in high-functioning adults with ASD.
Functional dissociations between different OXTR SNPs
Finally, we investigated biological functions of individual OXTR SNPs in terms of oxytocin’s effects in autism by examining the weights assigned to the SNPs in the regression analysis (e.g. if a SNP is assigned a positive regression weight, the SNP is supposed to enhance oxytocin efficacy).
As a result, we first found that all the weight values were significantly different from zero [t(3) > 7.6, PBonferroni-corrected < 0.05 in one-sample t-tests]. Notably, despite the independence between different types of oxytocin effects, the weight directions were the same across the different oxytocin efficacy (Figure 3D, Table 2), providing biological validation to the current observations.
Table 2.
Effects of OXTR SNPs on oxytocin efficacy
|
Effects of major alleles on oxytocin efficacy: increase (+) or decrease (–) |
|||||||
|---|---|---|---|---|---|---|---|
| SNP | Major allele | Nonverbal communication score | Response smoothness | ACC activity | dmPFC activity | dmPFC–ACC connectivity | NAA level in ACC |
| rs4686300 | CC | + | |||||
| rs237877/rs237878 | CC/TT | – | |||||
| rs11706648 | AA | + | + | + | + | ||
| rs237887 | GG | – | |||||
| rs2254295/rs2254298/rs2268491 | TT/GG/CC | – | – | – | – | – | |
| rs11131149 | GG | – | – | ||||
| rs53576 | GG | + | + | + | + | + | + |
| rs2268495/rs4686302 | GG/CC | – | – | – | |||
| rs237900 | GG | + | |||||
| rs17049528/rs6777726 | GG/GG | + | + | ||||
ACC, anterior cingulate cortex; dmPFC, dorsal medial prefrontal cortex; NAA, N-acetylaspartate.
In particular, the weights assigned to rs53576 were consistently positive in all the regressions for every oxytocin response, whereas weights for rs2254298 were always negative. These results suggest that individuals with the major allele type (GG) in rs53576 were generally more responsive to oxytocin than those with the minor alleles (GA/AA), whilst individuals with the minor alleles for this SNP (GA/AA) are more responsive to the neuropeptide in both behavioral and neural manners than those with the major allele (GG).
This distinct difference in the effects on oxytocin responsiveness between these two prominent OXTR SNPs was also confirmed by conventional univariate analyses [F(1, 221) > 6.9, P < 0.009 in repeated-measures two-way ANOVA; t(226) > 2.7, P < 0.008 in posthoc two-sample t-tests; Figure 4]. These results imply dissociable biological functions of different OXTR SNPs when oxytocin affects autistic social symptoms (see ‘Discussions’ section).
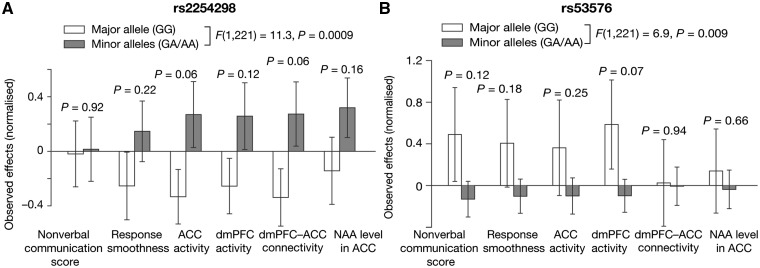
Fig. 4.
Comparison of oxytocin efficacy based on variations of a specific SNP. For the two prominent OXTR SNPs ((A) rs2254298 and (B) rs53576), we also performed univariate simple comparisons of the oxytocin efficacy between individuals with major allele and those with minor alleles. The overall trends that were evaluated as a main effect of SNP patters support the results of the original multivariate analyses [F(1, 221) > 6.9, P < 0.009 in two-way ANOVA of normalized oxytocin responses: (major homozygous allele, combined minor alleles) × (six types of oxytocin response)]. Error bars, SD.
Discussions
In this study, we identified specific OXTR SNP combinations that could robustly and accurately predict the magnitude of behavioral and neural effects of oxytocin on social impairments in Japanese high-functioning adult males with ASD. Moreover, these observations were qualitatively reproduced when we used the data recorded from 35 drug-free participants (Supplementary Materials, Supplementary Figures S7 and S8). Although these findings should be tested in a larger sample size in future studies, the current results provide new biological insights into the functions of several prominent OXTR variants, and this analysis procedure would contribute to establishment of future tailor-made precision medicines for ASD.
Biological implication
The current observations imply biological functions of several OXTR SNPs in ASD. In particular, the results summarized in Table 2 suggest seemingly distinct biological functions of two prominent OXTR SNPs, rs2254298 and rs53576, whose associations with ASD and their social behaviors have been repeatedly reported (Guastella et al., 2012; Kumsta and Heinrichs, 2013; Yamasue, 2013; Kanat et al., 2014). Although such associations may depend on ethnicity (Tost et al., 2011; Yamasue et al., 2011; Brüne, 2012), previous studies suggested that AA/AG allele types of rs2254298 and rs53576 (minor alleles for both SNPs) are relevant to atypical brain anatomy and relatively impaired social skills (Rodrigues et al., 2009; Inoue et al., 2010; Tost et al., 2010, 2011; Meyer-Lindenberg et al., 2011), and are more likely to be observed in individuals with ASD than in controls (Wu et al., 2005; Liu et al., 2010; Yamasue, 2013; LoParo and Waldman, 2015). Such relationships between specific OXTR SNP and impairment of social skills in ASD were also seen in the current dataset (Supplementary Figure S9): if baseline responses to the current social task are close to behavioral and neural responses to placebo, ASD individuals with AA/AG allele in rs2254298 and rs53576 showed more severely impaired social behaviors and less neural activity in the medial prefrontal cortex than those with GG allele [F(1,221) ≥ 7.3, P < 0.008 in repeated-measures two-way ANOVA].
Considering these previous findings and current observations, this study appear to indicate (i) that individuals with rs2254298A tend to have atypical neural systems for social cognition, show autistic symptoms, and be responsive to intranasal oxytocin administration, (ii) whilst those with rs53576A also tend to have atypical social cognition and be found in the autistic population, but they are likely to be less responsive to oxytocin than those with rs53576GG. This dissociation may also imply that, at least in the current Japanese participants, rs53576A might induce more severe functional impairments in oxytocin receptor than rs2254298A.
Comparison between the six markers
The adjusted R2s for neurobiological responses were larger than those for behaviors (Supplementary Table S2), and Akaike Information Criteria for these regressions (Supplementary Table S4) were lower in the neurobiological biomarkers than in behaviors. These comparisons suggest that genetic information more suitably fits to neural responses than to behavioral ones (Anagnostou et al., 2014), which is reasonable if we assume that genetic variations first induce neurobiological changes that thereby result in behavioral alterations. Given such high sensitive neural responses, drug trials might need to include neurobiological indices as their outcomes (Yamasue, 2015).
Clinical implication and significances
Clinically, the current results could help developments of precision medicines for ASD. In contrast to single-dose oxytocin administration (Guastella et al., 2009; Gordon et al., 2013; Domes et al., 2014; Aoki et al., 2014, 2015; Watanabe et al., 2014a), trials that continually administered oxytocin to ASD individuals appear to have experienced difficulty in detecting sufficient behavioral oxytocin effects (Anagnostou et al., 2012; Dadds et al., 2014; Guastella et al., 2015). This discrepancy may indicate that continual administration of oxytocin amplifies the individual variability in sensitivity to the neuropeptide. Therefore, for practical clinical application of oxytocin, it is increasingly necessary to identify personal characteristics that determine individual oxytocin efficacy. In this regard, our results suggest that, at least in the Japanese population, examining specific OXTR SNPs in Table 2 prior to actual oxytocin administration may help us to infer their oxytocin efficacy and to prevent ineffective pharmaceutical intervention.
Limitation
The generalization capability of the current results may be restricted by the participants’ ethnicity (i.e. Japanese). Previous studies suggested ethic differences in the ASD–OXTR associations (Brüne, 2012; Yamasue, 2013) and in relationships between OXTR variants and brain architecture (Tost et al., 2011; Yamasue et al., 2011). Therefore, we need to be cautious when generalizing the current observations to non-Asian groups.
Another limitation is the sample size. Previous pharmacogenetics studies shown some strong genome-wide associations in a relatively small sample size (Cirulli and Goldstein, 2010), and the current findings were robust against changes in analysis parameters and datasets. However, it is also the case that the current sample size may be insufficient for accurate regression analyses using a larger number of SNPs. Therefore, to examine more SNPs with sufficiently small inter-SNP multicollinearity, future studies will need to use larger sample.
Conclusion
We have examined associations between OXTR SNP variations and oxytocin efficacy in autism, and identified specific SNP sets that accurately predicted behavioral/neural responses to oxytocin in ASD. Although larger studies using different ethnic populations are necessary, the current findings and approach provide novel insight into neurobiological functions of ASD-related genetic variants, and would help the development of future precision medicines for ASD.
Funding
This study was supported by Grants-in-Aid for Scientific Research (KAKENHI) from the Japan Society for the Promotion of Science (Grant No. 26670535 to H.Y.); the Adaptable and Seamless Technology Transfer Program; the Centre of Innovation Program; CREST from Japan Science and Technology Agency; Medicinal-Grant from Takeda Science Foundation; and the Strategic Research Program for Brain Sciences by the Ministry of Education, Culture, Sports, Science, and Technology.
Conflict of interest. None declared.
Supplementary data
Supplementary data are available at SCAN online.
Supplementary Material
Acknowledgements
TW acknowledges the supports of the Japan Society for Promotion of Science and European Commission. We thank Prof Miho Kuroda for ASD diagnosis using Autism Diagnostic Observation Schedule.
References
- Anagnostou E., Soorya L., Brian J., et al. (2014). Intranasal oxytocin in the treatment of autism spectrum disorders: a review of literature and early safety and efficacy data in youth. Brain Research, 1580, 188–98. [DOI] [PubMed] [Google Scholar]
- Anagnostou E., Soorya L., Chaplin W., et al. (2012). Intranasal oxytocin versus placebo in the treatment of adults with autism spectrum disorders: a randomized controlled trial. Molecular Autism, 3, 16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y., Watanabe T., Abe O., et al. (2015). Oxytocin's neurochemical effects in the medial prefrontal cortex underlie recovery of task-specific brain activity in autism: a randomized controlled trial. Molecular Psychiatry, 20, 447–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y., Yahata N., Watanabe T., et al. (2014). Oxytocin improves behavioural and neural deficits in inferring others' social emotions in autism. Brain, 137, 3073–86. [DOI] [PubMed] [Google Scholar]
- Auyeung B., Lombardo M.V., Heinrichs M., et al. (2015). Oxytocin increases eye contact during a real-time, naturalistic social interaction in males with and without autism. Translation of Psychiatry, 5, e507.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop C.M. 2006. Pattern Recognition and Machine Learning. New York: Springer Verlag. [Google Scholar]
- Brüne M. (2012). Does the oxytocin receptor (OXTR) polymorphism (rs2254298) confer ‘vulnerability’ for psychopathology or “differential susceptibility?” Insights from evolution. BMC Medicine, 10, 38.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli E.T., Goldstein D.B. (2010). Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nature Reviews Genetics 11, 415–25. [DOI] [PubMed] [Google Scholar]
- Dadds M.R., MacDonald E., Cauchi A., Williams K., Levy F., Brennan J. (2014). Nasal oxytocin for social deficits in childhood autism: a randomized controlled trial. Journal of Autism and Developmental Disorders, 44, 521–31. [DOI] [PubMed] [Google Scholar]
- Devlin B., Scherer S.W. (2012). Genetic architecture in autism spectrum disorder. Current Opinion in Genetics and Development, 22, 229–37. [DOI] [PubMed] [Google Scholar]
- Drucker H., Burges C.J.C., Kaufman L., Smola A., Vapnik V.. 1997. Advances in Neural Information Processing Systems: Proceedings of the 1996 Conference 9, 155–161.
- Domes G., Kumbier E., Heinrichs M., Herpertz S.C. (2014). Oxytocin promotes facial emotion recognition and amygdala reactivity in adults with asperger syndrome. Neuropsychopharmacology, 39, 698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman D.J., Chen M.C., Gotlib I.H. (2011). Variant in oxytocin receptor gene is associated with amygdala volume. Psychoneuroendocrinology, 36, 891–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon I., Vander Wyk B.C., Bennett R.H., et al. (2013). Oxytocin enhances brain function in children with autism. Proceedings of the National Academy of Sciences of the United States of America, 110, 20953–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guastella A.J., Einfeld S.L., Gray K.M., et al. (2010). Intranasal oxytocin improves emotion recognition for youth with autism spectrum disorders. Biological Psychiatry, 67, 692–4. [DOI] [PubMed] [Google Scholar]
- Guastella A.J., Graustella A.J., MacLeod C. (2012). A critical review of the influence of oxytocin nasal spray on social cognition in humans: evidence and future directions. Hormones and Behavior, 61, 410–8. [DOI] [PubMed] [Google Scholar]
- Guastella A.J., Gray K.M., Rinehart N.J., et al. (2015). The effects of a course of intranasal oxytocin on social behaviors in youth diagnosed with autism spectrum disorders: a randomized controlled trial. Journal of Child Psychology and Psychiatry, 56, 444–52. [DOI] [PubMed] [Google Scholar]
- Guastella A.J., Howard A.L., Dadds M.R., Mitchell P., Carson D.S. (2009). A randomized controlled trial of intranasal oxytocin as an adjunct to exposure therapy for social anxiety disorder. Psychoneuroendocrinology, 34, 917–23. [DOI] [PubMed] [Google Scholar]
- Hinton G., Salakhutdinov R. (2006). Reducing the dimensionality of data with neural networks. Science, 313, 504–7. [DOI] [PubMed] [Google Scholar]
- Hollander E., Bartz J., Chaplin W., et al. (2007). Oxytocin increases retention of social cognition in autism. Biological Psychiatry, 61, 498–503. [DOI] [PubMed] [Google Scholar]
- Inoue H., Yamasue H., Tochigi M., et al. (2010). Association between the oxytocin receptor gene and amygdalar volume in healthy adults. Biological Psychiatry, 68, 1066–72. [DOI] [PubMed] [Google Scholar]
- Jacob S., Brune C.W., Carter C.S., Leventhal B.L., Lord C., Cook E.H. (2007). Association of the oxytocin receptor gene (OXTR) in Caucasian children and adolescents with autism. Neuroscience Letters 417, 6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.D., McDuff S.G.R., Rugg M.D., Norman K.A. (2009). Recollection, familiarity, and cortical reinstatement: a multivoxel pattern analysis. Neuron, 63, 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanat M., Heinrichs M., Domes G. (2014). Oxytocin and the social brain: neural mechanisms and perspectives in human research. Brain Research, 1580, 160–71. doi:10.1016/j.brainres.2013.11.003 [DOI] [PubMed] [Google Scholar]
- Kumar T.K. (1975). Multicollinearity in regression analysis. The Review of Economics and Statistics, 57, 365–6. [Google Scholar]
- Kumsta R., Heinrichs M. (2013). Oxytocin, stress and social behavior: neurogenetics of the human oxytocin system. Current Opinion in Neurobiology, 23, 11–6. [DOI] [PubMed] [Google Scholar]
- Lai M.C., Lombardo M.V., Baron-Cohen S. (2014). Autism. Lancet, 383, 896–910. [DOI] [PubMed] [Google Scholar]
- Lerer E., Levi S., Salomon S., Darvasi A., Yirmiya N., Ebstein R.P. (2008). Association between the oxytocin receptor (OXTR) gene and autism: relationship to Vineland Adaptive Behavior Scales and cognition. Molecular Psychiatry, 13, 980–8. [DOI] [PubMed] [Google Scholar]
- Li M.X., Kwan J.S.H., Sham P.C. (2012). HYST: a hybrid set-based test for genome-wide association studies, with application to protein-protein interaction-based association analysis. The American Journal of Human Genetics, 91, 478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Kawamura Y., Shimada T., et al. (2010). Association of the oxytocin receptor (OXTR) gene polymorphisms with autism spectrum disorder (ASD) in the Japanese population. Journal of Human Genetics, 55, 137–41. [DOI] [PubMed] [Google Scholar]
- Long N., Gianola D., Rosa G.J.M., Weigel K.A. (2011). Application of support vector regression to genome-assisted prediction of quantitative traits. Theoretical and Applied Genetics, 123, 1065–74. [DOI] [PubMed] [Google Scholar]
- LoParo D., Waldman I.D. (2015). The oxytocin receptor gene (OXTR) is associated with autism spectrum disorder: a meta-analysis. Molecular Psychiatry, 20, 640–6. [DOI] [PubMed] [Google Scholar]
- Lord C., Rutter M., Goode S., et al. (1989). Autism diagnostic observation schedule: a standardized observation of communicative and social behavior. Journal of Autism and Developmental Disorders, 19, 185–212. [DOI] [PubMed] [Google Scholar]
- Lord C., Rutter M., Le Couteur A. (1994). Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. Journal of Autism and Developmental Disorders, 24, 659–85. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A., Domes G., Kirsch P., Heinrichs M. (2011). Oxytocin and vasopressin in the human brain: social neuropeptides for translational medicine. Nature Reviews Neuroscience, 12, 524–38. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A., Tost H. (2012). Neural mechanisms of social risk for psychiatric disorders. Nature Neuroscience, 15, 663–8. [DOI] [PubMed] [Google Scholar]
- Miao-Xin Li H.S.G.J.S.H.K.P.C.S. (2011). GATES: a rapid and powerful gene-based association test using extended simes procedure. American Journal of Human Genetics, 88, 283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag C., Sauer C., Reuter M., Kirsch P. (2013). An interaction between oxytocin and a genetic variation of the oxytocin receptor modulates amygdala activity toward direct gaze: evidence from a pharmacological imaging genetics study. European Archives of Psychiatry and Clinical Neurosciences, 263(Suppl 2), S169–75. [DOI] [PubMed] [Google Scholar]
- Rodrigues S.M., Saslow L.R., Garcia N., John O.P., Keltner D. (2009). Oxytocin receptor genetic variation relates to empathy and stress reactivity in humans. Proceedings of the National Academy of Sciences of the United States of America, 106, 21437–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H. (2015). Neuroscience: The hard science of oxytocin. Nature, 522, 410–2. [DOI] [PubMed] [Google Scholar]
- Tost H., Kolachana B., Hakimi S., et al. (2010). A common allele in the oxytocin receptor gene (OXTR) impacts prosocial temperament and human hypothalamic-limbic structure and function. Proceedings of the National Academy of Sciences of the United States of America, 107, 13936–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tost H., Kolachana B., Verchinski B.A., et al. (2011). Neurogenetic effects of OXTR rs2254298 in the extended limbic system of healthy caucasian adults. Biological Psychiatry, 70, E37–9. [DOI] [PubMed] [Google Scholar]
- Thissen U., Pepers M., Üstün B., Melssen W.J., Buydens L.M.C. (2004). Chemometrics and Intelligent Laboratory Systems, 73, 169–79. [Google Scholar]
- Watanabe T., Abe O., Kuwabara H., et al. (2014a). Mitigation of sociocommunicational deficits of autism through oxytocin-induced recovery of medial prefrontal activity: a randomized trial. JAMA Psychiatry, 71, 166–75. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Hirose S., Wada H., et al. (2011). Prediction of subsequent recognition performance using brain activity in the medial temporal lobe. NeuroImage, 54, 3085–92. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Kuroda M., Kuwabara H., et al. (2015). Clinical and neural effects of six-week administration of oxytocin on core symptoms of autism. Brain, 138, 3400–12. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Yahata N., Abe O., et al. (2012). Diminished medial prefrontal activity behind autistic social judgments of incongruent information. PLoS One, 7, e39561.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Yahata N., Kawakubo Y., et al. (2014b). Network structure underlying resolution of conflicting non-verbal and verbal social information. Social Cognitive and Affective Neuroscience, 9, 767–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wermter A.K., Kamp-Becker I., Hesse P., Schulte-Körne G., Strauch K., Remschmidt H. (2010). Evidence for the involvement of genetic variation in the oxytocin receptor gene (OXTR) in the etiology of autistic disorders on high-functioning level. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 153B, 629–39. [DOI] [PubMed] [Google Scholar]
- Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. (2011). Rare-variant association testing for sequencing data with the sequence kernel association test. American Journal of Human Genetics, 89, 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Jia M., Ruan Y., et al. (2005). Positive association of the oxytocin receptor gene (OXTR) with autism in the Chinese Han population. Biological Psychiatry, 58, 74–7. [DOI] [PubMed] [Google Scholar]
- Yamasue H. (2015). Promising evidence and remaining issues regarding the clinical application of oxytocin in autism spectrum disorders. Psychiatry and Clinical Neurosciences [DOI] [PubMed] [Google Scholar]
- Yamasue H. (2013). Function and structure in social brain regions can link oxytocin-receptor genes with autistic social behavior. Brain Development, 35, 111–8. [DOI] [PubMed] [Google Scholar]
- Yamasue H., Suga M., Yahata N., et al. (2011). Reply to: neurogenetic effects of OXTR rs2254298 in the extended limbic system of healthy Caucasian adults. Biological Psychiatry, 70, 89–99. [DOI] [PubMed] [Google Scholar]
- Yatawara C.J., Einfeld S.L., Hickie I.B., Davenport T.A., Guastella A.J. (2016). The effect of oxytocin nasal spray on social interaction deficits observed in young children with autism: a randomized clinical crossover trial. Molecular Psychiatry, 21, 1225–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylisaukko-oja T., Alarcón M., Cantor R.M., et al. (2006). Search for autism loci by combined analysis of Autism Genetic Resource Exchange and Finnish families. Annals of Neurology, 59, 145–55. [DOI] [PubMed] [Google Scholar]
- Yrigollen C.M., Han S.S., Kochetkova A., et al. (2008). Genes controlling affiliative behavior as candidate genes for autism. Biological Psychiatry, 63, 911–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.