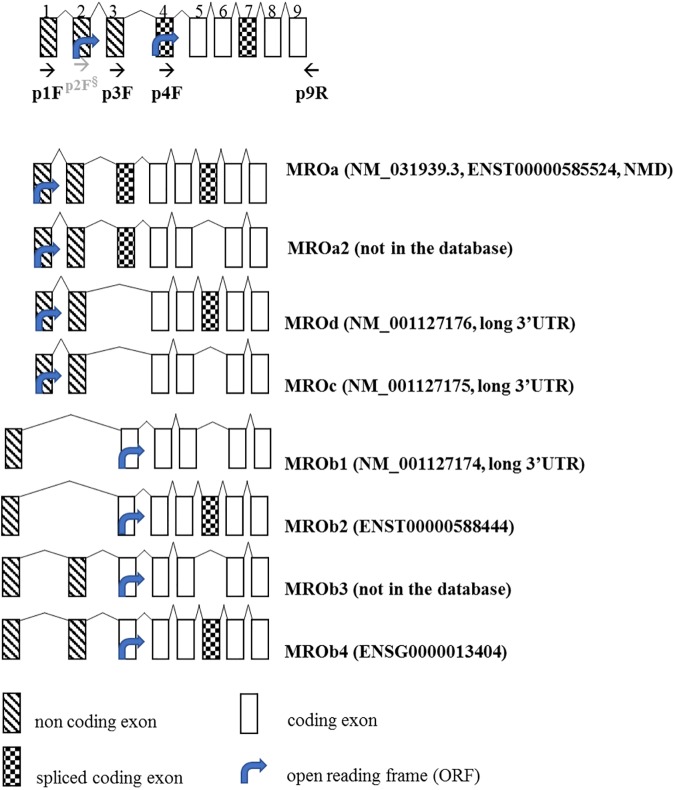
Fig 1. Schematic representation of the human Maestro (MRO) gene structure.
The start codon of the Open Reading Frame (ORF) indicated by an arrow is localized in exon 2 for MROa, MROa2, MROc, MROd, and in exon 4 for ‘MROb’ (b1-b4) transcript variants. MROa (NM_031939.3), MROd (NM_001127176.1), and MROc (NM_001127175.1) coding regions consist of 753bp, 648bp, and 492bp and encode 248aa, 262aa, and 210aa protein, respectively. MROb1 (NM_001127174), MROb2, and MROb3 consist of 678bp, 827bp, and 792bp and encode 196aa, 248aa, and 210aa proteins, respectively. MROa is considered as a non-sense mediated decay (NMD) transcript. The ‘a’ variant differs by a 103bp additional 5’ exon (exon 4) compared to the ‘c’ and ‘d’ variants. The ‘c’ variant lacks one in-frame exon (exon 7) compared to ‘d’. The MRO ‘b’ variants (MROb1-4) contain a distinct 5' UTR differ from MROa, c, d by a 76bp non-coding exon (exon 1), and differ from each other by one alternate exon (exon 3) and one in-frame exon (exon 7). MROb4 (BC029860.1) is the longest transcript, consisting of 948bp and encoding 248aa. The predictive protein isoforms have 2 distinct N-terminus with expected sizes of 196 aa– 248 aa and a mass of 23-29kDda (S1 Fig).