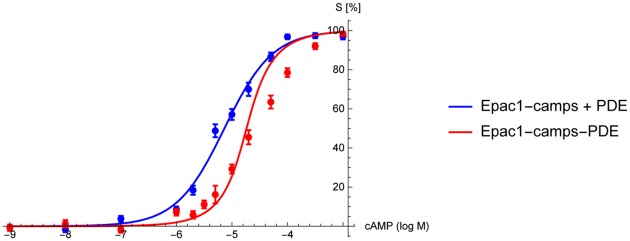
Fig 5. Binding curves of the free sensor protein Epac1-camps + PDE (blue) and the fusion protein Epac1-camps-PDE4A1 (red).
The overall PDE activity is equal in both experiments, indicating that the right shift of the binding curve is solely caused by local PDE degradation. The curves were determined by fitting Eq (18) to the corresponding experimental data. The distance between sensor and absorbing enzyme was set to d = 0nm as for an ideal sensor-enzyme construct. The fit of the free sensor data (blue) yields the dissociation constant of the sensor KD = 7.3 ± 0.66μM (goodness of fit: χ2 = 3.2, p = 0.66). From the sensor-enzyme construct (red) we obtain the absorptive action of a PDE η2 = 6.1 ± 1.4, (χ2 = 9.6, p = 0.08). The flooding of the nanocompartment as predicted by our model could not be observed in the experiments.