Abstract
Protein–protein interaction (PPI) and host–pathogen interactions (HPI) proteomic analysis has been successfully practiced for potential drug target identification in pathogenic infections. In this research, we attempted to identify new drug target based on PPI and HPI computation approaches and subsequently design new drug against devastating enterohemorrhagic Escherichia coli O104:H4 C277-11 (Broad), which causes life-threatening food borne disease outbreak in Germany and other countries in Europe in 2011. Our systematic in silico analysis on PPI and HPI of E. coli O104:H4 was able to identify bacterial d-galactose-binding periplasmic and UDP-N-acetylglucosamine 1-carboxyvinyltransferase as attractive candidates for new drug targets. Furthermore, computational three-dimensional structure modeling and subsequent molecular docking finally proposed [3-(5-Amino-7-Hydroxy-[1,2,3]Triazolo[4,5-d]Pyrimidin-2-Yl)-N-(3,5-Dichlorobenzyl)-Benzamide)] and (6-amino-2-[(1-naphthylmethyl)amino]-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one) as promising candidate drugs for further evaluation and development for E. coli O104:H4 mediated diseases. Identification of new drug target would be of great utility for humanity as the demand for designing new drugs to fight infections is increasing due to the developing resistance and side effects of current treatments. This research provided the basis for computer aided drug design which might be useful for new drug target identification and subsequent drug design for other infectious organisms.
Electronic supplementary material
The online version of this article (doi:10.1007/s40203-017-0021-5) contains supplementary material, which is available to authorized users.
Keywords: Drug resistance, Host–pathogen interactions, Homology modeling, Molecular docking, Protein structure
Background
Enteric bacterial infections remain a major cause of morbidity and mortality in both developing and developed countries (Petri et al. 2008). Among various pathogenic E. coli strains that cause intestinal or extra-intestinal diseases in humans, the most devastating are enterohemorrhagic E. coli (EHEC) strains, which produce highly potent cytotoxins called Shiga toxins (Stxs) (Bradley et al. 2012; Brooks et al. 2005). The EHEC E. coli caused diarrhea, hemorrhagic colitis, life-threatening hemolytic uremic syndrome and encephalopathy (Evans and Evans 1996). Several deadly EHEC outbreaks were reported all over the world since last two hundred years predominated by the O157:H7 strain of E. coli (Frank et al. 2011; King et al. 2012; Michino et al. 1999; Waters et al. 1994). However, in 2011, a new EHEC strain O104:H4 was identified and this strain was related to an outbreak in Germany and other countries in Europe. This massive outbreak was of great concern to the drug designers as a number of deaths from the infection were reported due to the ineffective medication (Bielaszewska et al. 2011; Grad et al. 2012). This pathogenic strain acquired a shiga toxin gene and an antibiotic resistant virulence plasmid pAA, which allow it to exhibit resistance against a significant number of antibiotics including cephalosporins, co-trimoxazole and all penicillins while susceptible to imipenem, meropenem, amikacin, kanamycin and carbapenems (Mora et al. 2011; Muniesa et al. 2012). In order to get the most effective treatment options, it is crucial to identify new drug and vaccine candidates to combat with this deadly pathogen.
The crucial step in drug discovery is the target identification (Chan et al. 2010). However, traditional drug discovery methods are capital-intensive, time-consuming, and often yield few drug targets. In contrast, advantages of the bioinformatics, genomics and proteomics approach represent an attractive alternative to identify drug targets worthy of experimental follow-up. The pathogen and host-genome sequence offer a better understanding of pathomechanism of disease and hence identification of drug targets. In recent years, computational methods have been used widely for the identification of potential drug and vaccine targets in different pathogenic microorganisms (Amineni et al. 2010; Damte et al. 2013; Mondal et al. 2014, 2015; Sliwoski et al. 2014). Protein–protein interactions (PPI) and host–pathogen interactions (HPI) approaches offer an area of unexplored potential for next generation drug targets (Taylor et al. 2011). It is important for bacterial cellular processes and pathogenesis analysis and thus efficient to identify the protein-set essential for the pathogen’s survival but absent in the host (Archakov et al. 2003; Eisenberg et al. 2000). Subtraction of the host genome from essential genes of pathogens helps in searching for non-human homologous targets which ensures no interaction of drugs with human targets. The integration of these approaches with different advanced bioinformatics tools may ensure the discovery of potential drug targets for most of the infectious diseases. After the drug target(s) optimization, the in silico virtual screening of different chemical databases could provide unprecedented opportunity to select and design the best possible inhibitor(s) (Lavecchia and Di Giovanni 2013).
This study focused on a combination approach of the proteomics data analysis and homology modeling to find out a novel therapeutic target from E. coli O104:H4 C277-11 (Broad). We performed the protein–protein interactions of E. coli O104:H4 through the three different methods (1) protein interactions from PSIbase (2) protein interactions from Database of Interacting Proteins (DIP) and (3) domain–domain interactions from Domain interaction map (DIMA). Host–pathogen interactions (HPIs) between predator E. coli O104:H4 and its target Homo sapiens were predicted by host–pathogen interaction database (HPIDB). E. coli O104:H4 proteins contributed in HPIs were investigated for identifying potential drug targets and subsequent computer aided drug design process. The identified potential drug targets might expand our understanding of the molecular mechanisms of E. coli O104:H4 pathogenesis and also facilitate the design of effective antibiotics.
Materials and methods
Intra species protein–protein interaction prediction from PSIbase, DIP and DIMA
Protein sequences of the E. coli O104:H4 str. C227-11 (Broad) was taken from the Patric Pathosystems Resource Integration Center (Gillespie et al. 2011). These sequences were assigned with Structural Classification of Proteins (SCOP) 1.75 database using SUPER FAMILY 1.75 with an e-value cutoff 0.0001 (de Lima Morais et al. 2011). SCOP domains were further submitted to the PSIbase database which is based on Protein Structural Interactome map (PSIMAP) information to obtain interaction partners (Gong et al. 2005b). A comparison of the protein sequences of E. coli O104:H4 str. C227-11 (Broad) to the protein sequences (dip20120218.seq) from DIP (Salwinski et al. 2004) database was carried out using BLASTp with an e value cutoff 10−5 and a minimum sequence identity of 40% (Altschul et al. 1997). Interaction pairs were obtained from the DIP database by applying DIP node identity from the blast output. All the E. coli O104:H4 proteins were aligned for the corresponding Pfam domains using WebMGA server with hmmscan 3.0 and PFAM 2.4 by the e value cutoff 0.01 (Luo et al. 2011; Wu et al. 2011).
Host–pathogen interactions from HPIDB
Homologous protein interactions between E. coli O104:H4 and human were predicted from the HPIDB (Kumar and Nanduri 2010). “Search Homologous HPIs” option was selected to identify host–pathogen interaction with the BLASTp parameter set to <1020 and the sequence identity cutoff of 30%.
Drug target identification
Pathogen proteins participating in host–pathogen interactions were further investigated for drug target identification.
Paralogous and non-homologous protein identification
The complete protein sequence of Homo sapiens was retrieved from NCBI FTP site (ftp://ftp.ncbi.nlm.nih.gov/). To identify duplicate or paralogous proteins within the 1497 proteins of E. coli O104:H4, protein sequences were purged at 60% using CD-HIT (Huang et al. 2010). Paralogs and duplicate proteins were discarded. The non-paralogous proteins were applied to BLASTp search against complete proteome of Homo sapiens to identify non-homologous proteins. Expectation threshold value was set to 10−4. The resultant dataset that had significant similarity with the human proteins were excluded and the non-homologous proteins were compiled.
Essential protein identification and metabolic pathway analysis
Non-homologous proteins were subjected to identify pathogen specific essential proteins. From the Database of Essential Genes (DEG) (Zhang et al. 2004) prokaryotes essential genes were downloaded and all of the non-homologous proteins were subjected to BLASTp against DEG database (de Lima Morais et al. 2011). Parameters set for BLASTp execution was cutoff for e value <10−10, bit score >100 and percentage of identity >35%. Metabolic pathway analysis of identifying essential proteins was carried out using KEGG Automatic Annotation Server (KAAS) (Moriya et al. 2007). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database was used for the comparative pathway analysis between E. coli O104:H4 and Homo sapiens to map out essential proteins entailed in pathogen specific metabolic pathway (Kanehisa and Goto 2000).
Subcellular localization prediction
Subcellular Localization prediction server PSORTb version 3.0.2 was used for the identification of cytoplasmic, periplasmic and transmembrane protein within the essential proteins involved in pathogen specific metabolic pathway (Yu et al. 2010).
Homology modeling, structure refinement and active site prediction
Homology modeling of d-galactose-binding periplasmic protein MglB and UDP-N-acetylglucosamine 1-carboxyvinyltransferase was performed by automated homology modeling server SWISS-MODEL using the templates retrieved from Protein Data Bank (PDB) by comparative modeling (Rajender et al. 2011). Three dimensional structure of d-galactose-binding periplasmic protein MglB and UDP-N-acetylglucosamine 1-carboxyvinyltransferase from the homology modeling were passed through the structure refinement process using KoBaMIN (Rodrigues et al. 2012). Structural Analysis and Verification Server (SAVES) was implemented for evaluating the quality and validation of the refined 3-D structure model (Luthy et al. 1992).
Active site in the validated refined model was predicted by the Computed Atlas of Surface Topography of proteins (CASTp) database (Dundas et al. 2006) with the default parameter. Through extensive literature search best active site was identified and selected for both of the models.
Virtual screening and docking ligand into homology model
Virtual screening was performed for identifying active lead compounds with total 6460 molecules, 1447 approved and 5040 experimental posited in DrugBank (Knox et al. 2011). All approved and experimental molecules are downloaded from DrugBank in SDF format and converted into mol2 format using OpenBabel 2.3.1 (O’Boyle et al. 2011). Raccoon v1.0 processed these mol2 formatted ligand file into Autodock Vina (Trott and Olson 2010) compatible format.
Cluster computer with Linux based with 32 core system was executed for virtual screening using mpiVina. This cluster computer capable to accomplish 500 docking per hour. Based on high binding affinity a simple python script selected top 100 molecules from the processed result. Re-docking of these 100 molecules was carried out several times to check the stable high binding affinity. Finally, Pymol, Discovery Studio was used to analyze and visualization of the ligand-receptor interaction.
ADMET pharmacokinetic and toxicological analysis
Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) analyses constitute the pharmacokinetics of a drug molecule. The PreADMET server (https://preadmet.bmdrc.org) was used to predict the ADME profiles and carcinogenicity of a compound and helps to avoid toxic compound.
Results and discussion
Analysis of E. coli O104:H4 C227-11 protein–protein interactions (PPI) and host–pathogen interaction PPI
A total of 5325 proteins of E. coli O104:H4 C227-11 (Broad) was used for protein–protein interaction analysis. The algorithm PSIMAP was reported to extrapolate the protein–protein interactions by applying the sequence similarities (Kim et al. 2008). It provides a global interactive map of protein–protein and domain–domain interaction information by calculating euclidean distance between structural domains in interacting protein pairs (Gong et al. 2005b). PSIMAP utilizes Protein Data Bank (PDB) and Structural Classification of Proteins (SCOP) domains to retrieve molecular interaction data and test the fundamental working mechanisms of cells (Gong et al. 2005a). We carried out E. coli O104:H4 protein interactions using SUPER FAMILY 1.75 and PSIbase. Our analysis identified 7769 predictive interactions for 1278 proteins which was around 24% of the total E. coli O104:H4 C227-11 (Broad) proteins (Supplementary file 1). On contrary, Database of Interacting Proteins (DIP) analysis revealed 17,411 protein–protein interactions (PPI) from 2512 proteins which was 47% of the total E. coli proteins (Supplementary file 2). All these PPI has experimental evidence because DIP deposit and unionize information on protein–protein interaction was extracted from single research articles (Salwinski et al. 2004).
Domain interaction map (DIMA) imported domain–domain interaction pairs from iPfam and 3DID database which have been known to build physical contacts to the Protein Data Bank (PDB) data (Luo et al. 2011). We predicted 136885 Pfam domain interaction pairs for 3451 proteins comprising 64% of the total proteins using DIMA. We predicted 11107 protein–protein interactions from iPfam composed of 2527 E. Coli proteins representing approximately 47% of the total proteins (Supplementary file 3).
To explore the infection strategies, host–pathogen protein interactions information is crucial. For this purpose, we analyzed the host–pathogen interactions of pathogenic E. coli O104:H4 C227-11 (Broad) with human. In this analysis, we detected 1493 E. coli proteins targeting 1910 human proteins and the total number of protein–protein interactions was 4657. The HPIDB server was used to predict the host–pathogen interactions which infer homologous protein interactions from its integrated databases (Kumar and Nanduri 2010). HPIDB integrates Biomolecular Interaction Network Database (BIND) (Bader et al. 2003), Molecular INTeraction database (MINT) (Zanzoni et al. 2002), pathogen interaction gateway (PIG) (Driscoll et al. 2009), Gene Reference Into Function (GeneRIF), REACTOME (Matthews et al. 2009), INACT (Aranda et al. 2010) databases into one station and serve as a unified resource to investigation of host–pathogen interaction. From the HPIDB result we excluded the protein sequences less than 30% sequence identity for pruning the resultant dataset. According to the E. Coli–Homo sapiens interaction results we observed that the protein sequences of E. coli O104:H4 C227-11 (Broad) showed significant homology with the Bacillus anthracis, Yersinia pestis, Francisella tularensis subsp. tularensis SCHU S4, E. coli K-12, E. coli O157:H7. Detailed data for this full set of interactions is given in Supplementary file 4.
Drug target identification from the E. coli O104:H4 C227-11 (Broad)
For identifying putative drug targets against E. coli O104:H4 C227-11 (Broad), we analyzed 1493 E. coli proteins that can interact or target their human host proteins. For rationalizing protein dataset, paralogs or duplicate proteins were discarded using CD-HIT. We identified 56 duplicate protein sequences and omitted from the total list. Finally, 1437 protein sequences were subjected to a BLASTP program for deciphering non-human homologous sequences. Non-human homologous protein should be selected for drug target to preclude the cross binding of drug to the human body and for avoid the consequences of adverse drug side effects (Sakharkar et al. 2004). Total 797 non-human homologous proteins were further tested for identification essential gene using DEG database (Zhang and Lin 2009). For development of new promising and potential antimicrobial drugs it is the prerequisite first to identify essential genes that own fundamental role in the bacterial cellular process (Zhang and Lin 2009). From the BLASTP result, 339 essential proteins were identified which are crucial for the survival of E. coli O104:H4 C227-11 (Broad) bacterium. Metabolic pathway analysis of essential proteins was carried out by the KEGG Automatic Annotation server (KAAS) (Moriya et al. 2007). From 339 essential proteins 71 metabolic pathways were constructed and compared to the Homo sapiens metabolic pathways for drawing pathogen specific unique metabolic pathway. We select 17 unique pathways comprising 79 essential proteins to map out best candidate for drug targets by predicting their subcellular localization in a cell.
Computational anticipation of protein subcellular localization acts an important agent for detecting potential drug targets (Yu et al. 2010). Subcellular localization of essential proteins in unique pathways was accomplished by PSORTb (Yu et al. 2010). Out of 79 proteins, 49 proteins were found to be cytoplasmic, 25 to be localized in the cytoplasmic membrane, 1 outer membrane protein, 2 proteins found to be periplasmic localized and the remaining 2 proteins location were unknown. Although the membrane proteins, particularly receptor proteins and ion channels, have great importance in a wide variety of therapeutics, there are practical problems of working with membrane proteins like as protein purification, expression and crystallization (Duffield et al. 2010; Arinaminpathy et al. 2009). We selected best candidate by extensive literature review from cytoplasmic and periplasmic proteins. Finally, two potential drug targets are selected in Galactose/methyl galactoside ABC transport system, d-galactose-binding periplasmic protein MglB and UDP-N-acetylglucosamine 1-carboxyvinyltransferase (Fig. 1a, b). Drug target identification steps and their results are summarized (Table 1).
Fig. 1.
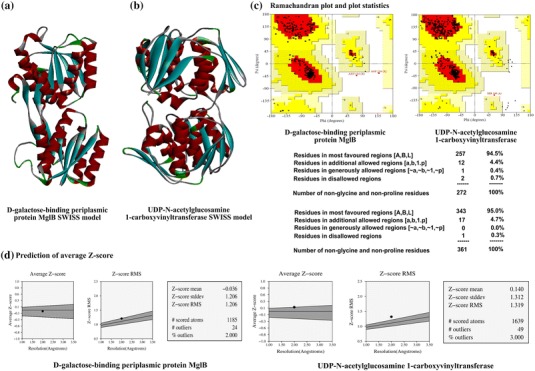
Homology modeling and structural quality check of drug target candidates from E. coli O104:H4 str. C227-11. Homology modeling by using SWISS-MODEL server based on template structure 2FVY and 3KQJ. a d-Galactose-binding periplasmic protein MglB, and b UDP-N-acetylglucosamine 1-carboxyvinyltransferase. c Structure quality validation by using the PROCHEK program. PROCHECK program executes Ramachandran plot and plot statistics for d-galactose-binding periplasmic protein MglB, and UDP-N-acetylglucosamine 1-carboxyvinyltransferase protein. d Prediction of average Z score of d-galactose-binding periplasmic protein MglB, and UDP-N-acetylglucosamine 1-carboxyvinyltransferase protein using PROVE
Table 1.
A summary of drug target identification steps and the results
| Drug target identification steps | Results |
|---|---|
| Total proteins | 1493 |
| Duplicate proteins identified by CD-HIT | 56 |
| Final set of proteins without duplicates or paralogs | 1437 |
| Non-homologous to human | 797 |
| Essential proteins identified from DEG | 339 |
| Metabolic pathway construction by essential proteins | 71 |
| Pathogen specific unique pathways | 17 |
| Essential proteins involved in unique pathways | 79 |
| Cytoplasmic proteins | 49 |
| Cytoplasmic membrane proteins | 25 |
| Outer membrane protein | 1 |
| Periplasmic proteins | 2 |
| Unknown localized proteins | 2 |
d-Galactose-binding periplasmic protein involved in the active transport of glucose and galactose. It mediates chemotaxis towards the two sugar residue by interacting with the Trg chemoreceptor in a number of bacterial species (Borrok et al. 2007). By chemotaxis bacteria encounter the environmental surrounding and find out more favorable conditions for its survival (Rao et al. 2004). d-Galactose-binding periplasmic protein plays a crucial role in the bacterial chemotaxis which emphasizes this protein as a potential drug target. UDP-N-acetylglucosamine 1-carboxyvinyltransferase, alternative name is UDP-N-acetylglucosamine enolpyruvyl transferase, participates for the biosynthesis of peptidoglycan polymer (Gautam et al. 2011; Kumar et al. 2009). Peptidoglycan serves as a crucial component of bacterial cell wall and important for bacterial survival. Furthermore, this essential enzyme is omnipresent in all the prokaryotes but not required by mammals. Therefore UDP-N-acetylglucosamine 1-carboxyvinyltransferase is an attractive target for the drug design to pursue against the drug resistant bacteria (Eschenburg et al. 2005).
Homology modeling, structure refinement and validation
Homology modeling was performed to determine the 3D-structure of d-galactose-binding periplasmic and UDP-N-acetylglucosamine 1-carboxyvinyltransferase. Template identification of the target proteins was carried out based on the sequence identities, lower expect value, X-ray crystallography resolution and R value. We selected 2FVY for d-galactose-binding periplasmic protein and 3KQJ for UDP-N-acetylglucosamine 1-carboxyvinyltransferase as the most suitable templates using the aforementioned criteria. The complete results of template identification for these two drug target proteins are presented (Table 2). Homology modeling was performed by using SWISS-MODEL server based on template structures 2FVY and 3KQJ, respectively (Fig. 1a, b).
Table 2.
Results of template identification for two drug target proteins based on the sequence identities, lower expect value, X-ray crystallography resolution and R value
| Drug target protein | Template PDB ID | Organism | % of identity | E value | X ray crystallography resolution (Å) | R value |
|---|---|---|---|---|---|---|
| d-Galactose-binding periplasmic | 2FVY | Escherichia coli | 95 | 1.30291E−167 | 0.92 | 0.118 |
| UDP-N-acetylglucosamine 1-carboxyvinyltransferase | 3KQJ | Escherichia coli K-12 | 97 | 0.0 | 1.70 | 0.168 |
Structure refinement for 3D homology model of d-galactose-binding periplasmic protein and UDP-N-acetylglucosamine 1-carboxyvinyltransferase was carried out by KoBaMIN web server (Rodrigues et al. 2012). To refine the protein structure after homology modeling KoBaMIN provides a fast and effective protein structure refinement based on minimization of a knowledge-based potential of mean force (Rodrigues et al. 2012). The initial energy of d-galactose-binding periplasmic homology structure was −3867.28 kcal/mol and after refinement final energy appeared to be −6706.5036 kcal/mol. Total energy minimizations was −2839.22 kcal/mol. In addition, UDP-N-acetylglucosamine 1-c−rboxyvinyltransferase homology structure energy minimization was −3918.85 kcal/mol (Fig. 1a, b; Supplementary file 5).
Further we determined the structure quality by validating the modeled PDB. We used SAVES metaserver (which integrates 6 programs PROCHECK, WHAT_CHECK, ERRAT, VERIFY_3D, PROVE, CRYST1 record matches) to check structural quality (http://services.mbi.ucla.edu/SAVES/) (Pontius et al. 1996). PROCHECK program executes the Ramachandran plot to evaluate model quality (Hariprasad et al. 2010). Ramachandran plot analysis of the 3D structures of d-galactose-binding periplasmic protein showed that 94.5% residues in the core region, 4.4% in allowed region. ERRAT and PROVE results for the same protein structure showed quality factor 96.633 and average Z score −0.036, respectively. On contrary, UDP-N-acetylglucosamine 1-carboxyvinyltransferase Ramachandran plot expressed that 95.0% of the residues are in the core region. (Figure 1c, d and Supplementary file 6).
Active site prediction and virtual screening for docking
Drug targets proteins active sites were predicted by the Computed Atlas of Surface Topography of proteins (CASTp). Using CASTp, we selected pocket volume 1006.7 and 1737.2 for d-galactose-binding periplasmic protein and UDP-N-acetylglucosamine 1-carboxyvinyltransferase proteins, respectively (Fig. 2). Comparative active site analysis of 2FVY (template) and d-galactose-binding periplasmic protein (homology model) reveal 100% conserved residues between template and homology model.
Fig. 2.
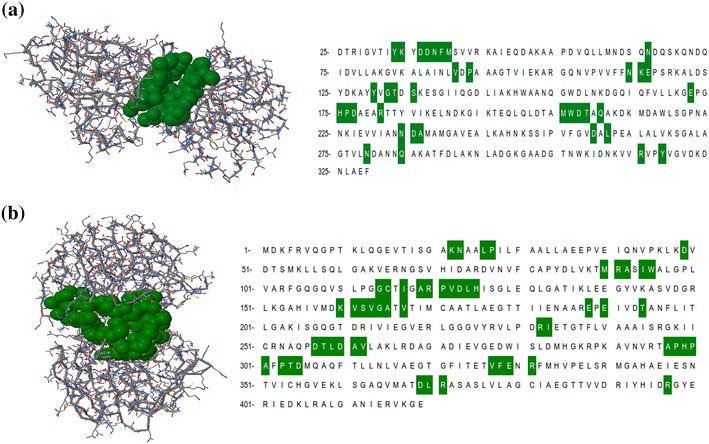
Active site residues of drug target candidates from E. coli O104:H4 str. C227-11. Identification of the active site was done by CASTp server. Two significant binding pockets of a d-galactose-binding periplasmic protein MglB, and b UDP-N-acetylglucosamine carboxyvinyltransferase protein. Amino acid residues of the respective binding pockets are indicated in green
Follow the prediction of the active site residues we carried out molecular docking using mpiVina based on lowest binding energy (Azam and Abbasi 2013). Top 100 ligands were selected from the result dataset which were showing lower binding energy to the receptor’s active site filtered for Lipinski’s rule of 5 (Lipinski et al. 2001). 81 ligands follow the Lipinski’s rule of 5 and out of 81 inhibitors top 10 ligands having lower binding energies were selected and most importantly bacterial proteins as their targets. Inhibitors that target human proteins were filtered out for avoiding off target binding with human proteins. Finally, top 10 hits for both of the target proteins d-galactose-binding periplasmic protein and UDP-N-acetylglucosamine 1-carboxyvinyltransferase protein were listed according to their binding energy. Top 10 docking results of these two target proteins are represented (Tables 3, 4).
Table 3.
Top ten docking results for d-galactose-binding periplasmic protein
| No. | Screened compounds (Drug Bank id) | Binding energy (kcal/mol) | Active site residues involved in interactions | Molecular weight (g/mol) |
|---|---|---|---|---|
| 1 | DB03571 | −10.5 | Gln 210, Asp 207, Thr 208, Asn 38, Asp 37, Ala 236, Phe 39, Asn 234, Asn 279, Asp 259, Asn 114, Arg 181, Asp 177, His 175, Glu 172, Trp 206, Tyr 33, Lys 34, Met 205, Asn 66 | 430.248 |
| 2. | DB06949 | −10.2 | Asn 114, His 175, Thr 208, Asp 207, Trp 206, Asp 37, Met 205, Lys 34, Tyr 33, Leu 261, Asn 38, Asn 234, Phe 39, Arg 181, Ala 236, Asp 259, Asn 279, | 464.107 |
| 3 | DB08666 | −10.2 | Tyr 318, Gln 284, Phe 39, Asn 279, Arg 181, Asp 259, Tyr 33, Asn 234, Asp 37, Trp 206, Asn 114, His 175, Asp 177, Tyr 130, Gly 132, Thr 133, | 268.198 |
| 4 | DB01691 | −10.1 | Gln 210, Asn 66, Lys 34, Glu 172, Lys 115, Asn 114, His 175, Asn 279, Arg 181, Asn 234, Asp 259, Trp 206, Asp 37, Tyr 33, Met 205, | 374.4357 |
| 5 | DB07055 | −10.1 | Trp 206, Asp 259, Asn 234, Asn 279, Gln 284, Arg 181, Tyr 130, Thr 133, Gly 132, Ser 135, Asp 177, Tyr 318, Asn 114, His 175, Asp 37, | 243.2215 |
| 6 | DB07434 | 9.8 | Asp 177, Asn 114, Arg 181, Asp 37, Thr 208, Tyr 33, Asp 36, Asp 207, Lys 34, Gln 210, Asn 66, Met 205, Asn 38, Ala 236, Asn 234, Trp 206, Phe 39, Asp 259, His 175 | 457.4366 |
| 7 | DB08668 | −9.8 | Gly 132, Tyr 130, Thr 133, Asp 177, Lys 115, His 175, Tyr 33, Trp 206, Asn 234, Arg 181, Asp 259, Asn 114, Gln 284, Phe 39, Asn 279, | 244.2095 |
| 8 | DB07041 | −9.7 | Phe 39, Asn 38, Thr 208, Ala 236, Asp 207, Asn 234, Lys 34, Trp 206, Met 205, Met 40, Asn 114, Glu 172, Lys 115, His 175, Asn 66, Tyr 33, Gln 210, Asp 36, Asp 37 | 428.4864 |
| 9 | DB07743 | −9.7 | Asn 234, Asn 279, Asn 66, Lys 115, Trp 206, Asn 38, Lys 34, Asp 37, Thr 208, Asp 36, Asp 207, His 175, Glu 172, Met 205, Tyr 33, Asn 114, Asp 259, Arg 181 | 363.314 |
| 10 | DB04452 | −9.5 | Asn 279, Arg 181, Asp 259, Phe 39, Asn 234, Asn 66, Trp 206, Asp 37, Lys 34, Tyr 33, Glu 172, Met 205, His 175, Asp 177, Asn 114 | 372.4231 |
Lowest docking energies, important residues of the binding site observed to be interactive with the ligands from DrugBank
Table 4.
Top ten docking results for UDP-N-acetylglucosamine 1-carboxyvinyltransferase
| No. | Screened compounds (Drug Bank Id) | Binding Energy (kcal/mol) | Active site residues involved in interactions | Molecular weight (g/mol) |
|---|---|---|---|---|
| 1 | DB04118 | −11.2 | Glu 190, Trp 95, Asn 23, Ala 165, Glu 188, Gly 164, Ser 162, His 125, His 299, Val 163, Pro 298, Val 161, Phe 328, Val 327, Glu 329, Arg 120, Arg 91, Arg 232 | 491.5372 |
| 2 | DB02033 | −10.9 | His 125, Arg 120, Ala 92, Arg 91, Phe 328, Leu 26, Pro 121, Lys 22, Thr 304, Asp 305, Val 163, Asn 23, Gly 164, Val 327, Trp 95, Ser 162 | 479.588 |
| 3 | DB01061 | −10.8 | Pro 121, Leu 124, Val 327, Phe 328, Pro 298, His 299, Glu 188, Asp 305, Arg 232, Thr 304, Asn 23, Val 163, Gly 164, Val 161, Ser 162, Ala 165, Arg 120, Arg 91, His 125 | 461.492 |
| 4 | DB08512 | −10.8 | Gly 398, Asp 49, Leu 370, Lys 22, Arg 371, Asn 23, Thr 304, Asp 305, Arg 232, Val 163, Pro 298, Glu 188, His 299, Val 327, Cys 115, Arg 120, Arg 91, Gly 114, Arg 397 | 356.3806 |
| 5 | DB01459 | −10.7 | Leu 370, Asp 49, Arg 397, Lys 22, Arg 91, Phe 328, Val 327, Pro 298, Val 163, Ser 162, Val 161, Gly 164, Thr 304, Asp 305, Asn 23, Arg 120, Trp 95,Ala 92, Cys 115 | 492.6114 |
| 6 | DB04030 | −10.7 | Val 327, Val 161, Pro 298, Ser 162, Val 163, His 125, Arg 91, Gly 164, Arg 120, Asn 23, Arg 232, Glu 190 | 398.4538 |
| 7 | DB04064 | −10.7 | Val 163, Trp 95, Asn 23, Asp 305, Arg 232, Glu 188, Thr 305, Pro 303, His 299, Ala 297, Pro 298, Val 327, Arg 91, Phe 328, Arg 120, Ser 162, Gly 164 | 378.3317 |
| 8 | DB04685 | −10.7 | Asp 49, Met 90, Arg 91, Arg 397, Ile 117, Gly 114, Cys 115, Trp 95, His 125, Ser 162, Gly 164, Val 163, Thr 304, Val 327, Asp 305, Asn 23, Arg 120, Phe 328, Lys 22, Leu 26, Ala 92 | 472.5075 |
| 9 | DB06922 | −10.7 | Asn 23, His 125, Arg 91, Ala 165, Gly 164, Arg 120, Ala 119, Glu 329, Val 327, Phe 328, Val 163, Ser 162 | 389.3576 |
| 10 | DB07241 | −10.7 | His 125, Arg 91, Lys 160, Arg 120, Ser 162, Val 327, Val 161, Val 163, Pro 298, Glu 188, His 299, Gly 164 | 355.3462 |
Lowest docking energies, important residues of the binding site observed to be interactive with the ligands from DrugBank
Virtual screening result for d-galactose-binding periplasmic protein shows that DB03571 [3-(5-Amino-7-Hydroxy-[1,2,3]Triazolo[4,5-d]Pyrimidin-2-Yl)-N-(3,5-Dichlorobenzyl)-Benzamide)] have high binding affinity to the protein with lowest binding energy −10.5 kcal/mol. This ligand was found to interact with Gln 210, Asp 207, Thr 208, Asn 38, Asp 37, Ala 236, Phe 39, Asn 234, Asn 279, Asp 259, Asn 114, Arg 181, Asp 177, His 175, Glu 172, Trp 206, Tyr 33, Lys 34, Met 205 and Asn 66. Docking study results for UDP-N-acetylglucosamine-1-carboxyvinyltransferase protein brings out that DB04118 (N-coeleneterazine) contact with target protein’s active site with high binding affinity (binding energy = −11.2 kcal/mol but among the top 10 hits we selected DB08512 (6-amino-2-[(1-naphthylmethyl)amino]-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one) having both high binding affinity (10.7 kcal/mol) and interaction with previously reported essential active site residues Cys 115, Asn 23, Asp 305 and Lys 22 (Fig. 3) (Samland et al. 1999). Blocking of these active site residues in UDP-N-acetylglucosamine 1carboxyvinyltransferase can inhibit the first committed step for peptidoglycan biosynthesis of bacterial cell. Therefore, the docking results and interactions with the essential active sites, suggested that DB03571 [3-(5-Amino-7-Hydroxy-[1,2,3]Triazolo[4,5-d]Pyrimidin-2-Yl)-N-(3,5-Dichlorobenzyl)-Benzamide)] and DB08512 (6-amino-2-[(1-naphthylmethyl)amino]-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one) might be used as potential inhibitors against the E. coli O104:H4 C227-11 (Broad) mediated diseases.
Fig. 3.
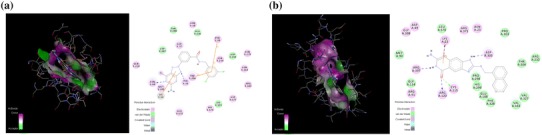
Molecular docking of predicted inhibitors to the binding site of a d-galactose-binding periplasmic protein MglB, and b UDP-N-acetylglucosamine 1-carboxyvinyltransferase protein are represent in 2D and 3D diagram. The color indicator on the left side of both diagrams indicates the types of interaction of particular residues
ADMET pharmacokinetic and toxicological analysis
Prediction and significant description of drug-likeness such as absorption, distribution, metabolism, excretion (ADME) and toxic (Tox) properties were calculated with the help of online server PreADMET. Through this server, we can calculate different parameters such as human intestinal absorption (HIA), cellular permeability (Pcaco-2), cell permeability Maden Darby Canine Kidney (PMDCK), plasma protein binding (PPB), carcinogenicity, and penetration of the blood–brain barrier (Cbrain/Cblood). The ADME properties should be perfect for a drug candidate. Both of our proposed drug (DB03571 and DB08512) shows proper ADME properties (Table 5).
Table 5.
The ADME and Toxicological analyses of DB03571 and DB08512 drug candidate compounds
| No. | Properties | DB03571 | DB08512 |
|---|---|---|---|
| 1 | HIA (%) | 94.07 | 88.61 |
| 2 | MDCK (nm/s) | 0.08 | 0.39 |
| 3 | Caco-2 (nm/s) | 19.85 | 20.84 |
| 4 | BBB (b/b) | 0.02 | 0.25 |
| 5 | PPB (%) | 92.26 | 86.92 |
| 6 | Carcinogenicity (rats) | Negative | Negative |
In general, HIA indicates ‘poor’ absorption in the range of 0–20%, ‘moderate’ absorption from 20 to 70% and ‘well’ absorption between 70 and 100%. We found HIA was 94.07% and 88.61% for DB03571 and DB08512, respectively, indicating well-absorbed compounds. The MDCK computational component would predict the renal clearance of the molecule. The MDCK results showed 0.08 nm/s and 0.39 nm/s for DB03571 and DB08512, respectively. Both compounds show very low permeability in in vitro MDCK cells (less than 25 nm/s according to PreADMET). The value of in vitro cell permeability (Pcaco-2) on intestinal epithelium cells are considered as ‘low’ permeability when the value is less than 4 nm/s, ‘middle’ permeability when the range is from 4 to 70 nm/s and high permeability when it is above 70 nm/s. The observed cell permeability of DB03571 and DB08512 are 19.85 and 20.84 nm/s, respectively, in intestinal epithelium and it indicates middle permeability in in vitro Caco-2 cells.
Blood brain barrier (BBB) restricts the passage of most of the compounds from the blood to brain, thus having a brain protecting property. The in vivo blood brain barrier (BBB) is the direct measure of penetration of drug in central nervous system (CNS). The DB03571 and DB08512 are found to be low and middle absorption category according to PreADMET analysis. Plasma protein binding prediction results showed 92.26 and 86.92% plasma protein binding for DB03571 and DB08512, respectively, indicating strongly bound chemicals which are not desirable.
Generally, the carcinogenicity test requires much study time (>2 years) in case of experimental procedures, but PreADMET helps to analyse the results quickly from NTP (National toxicology program) and EUA/FDA 2 year’s in vivo data. Both drugs exhibit no evidence of carcinogenic activity. So, the overall ADME properties of DB03571 and DB08512 are excellently satisfactory.
Conclusion
Through the integrated protein–protein interaction and host–pathogen interaction analysis, we identified two potential drug targets from the cytoplasmic and periplasmic region of E. coli 01O4:H4 C227-11 (Broad). The first target d-galactose-binding periplasmic protein play important role in bacterial chemotaxis. In chemotaxis, bacteria sense chemical gradients in the environment and move toward favorable conditions for survival. The pathway is arguably best characterized in the case of E. coli. Whereas UDP-N-acetylglucosamine 1-carboxyvinyltransferase target protein function in the peptidoglycan biosynthesis which is crucial for bacterial cell wall formation and thus important for bacterial survival. Most importantly, this enzyme is essential for bacteria but absent in human host. Extensive literature review supports both of this protein as prominent drug target candidate.
Bacteria become resistant against single or multiple antibiotics and ensure their survival in the environment. The identification of potential targets for the development of new antibiotics is becoming a topical and widely recognized need. The present study is an important basis for screening novel and alternative targets in a way to design and develop new drugs against other emerging human pathogens.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We acknowledge Professor Dr. Muhammed Zafar Iqbal, SUST for his cooperation and supportive hands throughout the research. We would like to thank Md. Mokkaram Hossain for his valuable support during blasting in cluster computer.
Abbreviations
- EHEC
Enterohemorrhagic E. coli
- HPI
Host–pathogen interactions
- PPI
Protein–protein interactions
- DIP
Database of Interacting Proteins
- DIMA
Domain interaction map
- HPIDB
Host–Pathogen Interaction Database
- SCOP
Structural Classification of Proteins
- PSIMAP
Protein Structural Interactome map
- DEG
Database of Essential Genes
- KAAS
KEGG Automatic Annotation Server
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PDB
Protein Data Bank
- SAVES
Structural Analysis and Verification Server
- CASTP
Computed Atlas of Surface Topography of proteins
- ADMET
Absorption, Distribution, Metabolism, Excretion and Toxicity
- HIA
Human intestinal absorption
- BBB
Blood brain barrier
Author contributions
SIM and ZM build the idea and wrote the manuscript. SIM, ZM and ME designed the study. SIM and ZM performed all the analysis and others helped in analysis. All authors read and approved the final manuscript.
Compliance with ethical standards
Competing interest
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s40203-017-0021-5) contains supplementary material, which is available to authorized users.
Shakhinur Islam Mondal, Zabed Mahmud and Montasir Elahi are equally contributed to this work.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amineni U, Pradhan D, Marisetty H. In silico identification of common putative drug targets in Leptospira interrogans. J Chem Biol. 2010;3(4):165–173. doi: 10.1007/s12154-010-0039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38(Database issue):D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archakov AI, Govorun VM, Dubanov AV, Ivanov YD, Veselovsky AV, Lewi P, et al. Protein–protein interactions as a target for drugs in proteomics. Proteomics. 2003;3(4):380–391. doi: 10.1002/pmic.200390053. [DOI] [PubMed] [Google Scholar]
- Arinaminpathy Y, Khurana E, Engelman DM, Gerstein MB. Computational analysis of membrane proteins: the largest class of drug targets. Drug Discov Today. 2009;14(23–24):1130–1135. doi: 10.1016/j.drudis.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam SS, Abbasi SW. Molecular docking studies for the identification of novel melatoninergic inhibitors for acetylserotonin-O-methyltransferase using different docking routines. Theor Biol Med Model. 2013;10:63. doi: 10.1186/1742-4682-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Betel D, Hogue CW. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31(1):248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielaszewska M, Mellmann A, Zhang W, Kock R, Fruth A, Bauwens A, et al. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis. 2011;11(9):671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- Borrok MJ, Kiessling LL, Forest KT. Conformational changes of glucose/galactose-binding protein illuminated by open, unliganded, and ultra-high-resolution ligand-bound structures. Protein Sci Publ Protein Soc. 2007;16(6):1032–1041. doi: 10.1110/ps.062707807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley KK, Williams JM, Burnsed LJ, Lytle MB, McDermott MD, Mody RK, et al. Epidemiology of a large restaurant-associated outbreak of Shiga toxin-producing Escherichia coli O111:NM. Epidemiol Infect. 2012;140(9):1644–1654. doi: 10.1017/S0950268811002329. [DOI] [PubMed] [Google Scholar]
- Brooks JT, Sowers EG, Wells JG, Greene KD, Griffin PM, Hoekstra RM, et al. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J Infect Dis. 2005;192(8):1422–1429. doi: 10.1086/466536. [DOI] [PubMed] [Google Scholar]
- Chan JN, Nislow C, Emili A. Recent advances and method development for drug target identification. Trends Pharmacol Sci. 2010;31(2):82–88. doi: 10.1016/j.tips.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Damte D, Suh JW, Lee SJ, Yohannes SB, Hossain MA, Park SC. Putative drug and vaccine target protein identification using comparative genomic analysis of KEGG annotated metabolic pathways of Mycoplasma hyopneumoniae. Genomics. 2013;102(1):47–56. doi: 10.1016/j.ygeno.2013.04.011. [DOI] [PubMed] [Google Scholar]
- de Lima Morais DA, Fang H, Rackham OJ, Wilson D, Pethica R, Chothia C, et al. SUPERFAMILY 1.75 including a domain-centric gene ontology method. Nucleic Acids Res. 2011;39(Database issue):D427–D434. doi: 10.1093/nar/gkq1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll T, Dyer MD, Murali TM, Sobral BW. PIG—the pathogen interaction gateway. Nucleic Acids Res. 2009;37(Database issue):D647–D650. doi: 10.1093/nar/gkn799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield M, Cooper I, McAlister E, Bayliss M, Ford D, Oyston P. Predicting conserved essential genes in bacteria: in silico identification of putative drug targets. Mol BioSyst. 2010;6(12):2482–2489. doi: 10.1039/c0mb00001a. [DOI] [PubMed] [Google Scholar]
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34(Web Server issue):W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Marcotte EM, Xenarios I, Yeates TO. Protein function in the post-genomic era. Nature. 2000;405(6788):823–826. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- Eschenburg S, Priestman M, Schonbrunn E. Evidence that the fosfomycin target Cys115 in UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) is essential for product release. J Biol Chem. 2005;280(5):3757–3763. doi: 10.1074/jbc.M411325200. [DOI] [PubMed] [Google Scholar]
- Evans DJ, Evans DG. Escherichia Coli in diarrheal disease. In: Baron S, editor. Medical microbiology. 4. Galveston, TX: University of Texas Medical Branch at Galveston; 1996. [PubMed] [Google Scholar]
- Frank C, Werber D, Cramer JP, Askar M, Faber M, an der Heiden M, et al. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med. 2011;365(19):1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- Gautam A, Rishi P, Tewari R. UDP-N-acetylglucosamine enolpyruvyl transferase as a potential target for antibacterial chemotherapy: recent developments. Appl Microbiol Biotechnol. 2011;92(2):211–225. doi: 10.1007/s00253-011-3512-z. [DOI] [PubMed] [Google Scholar]
- Gillespie JJ, Wattam AR, Cammer SA, Gabbard JL, Shukla MP, Dalay O, et al. PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infect Immun. 2011;79(11):4286–4298. doi: 10.1128/IAI.00207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Yoon G, Jang I, Bolser D, Dafas P, Schroeder M, et al. PSIbase: a database of Protein Structural Interactome map (PSIMAP) Bioinformatics. 2005;21(10):2541–2543. doi: 10.1093/bioinformatics/bti366. [DOI] [PubMed] [Google Scholar]
- Gong S, Park C, Choi H, Ko J, Jang I, Lee J, et al. A protein domain interaction interface database: InterPare. BMC Bioinform. 2005;6:207. doi: 10.1186/1471-2105-6-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad YH, Lipsitch M, Feldgarden M, Arachchi HM, Cerqueira GC, Fitzgerald M, et al. Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc Natl Acad Sci USA. 2012;109(8):3065–3070. doi: 10.1073/pnas.1121491109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariprasad G, Kumar M, Kaur P, Singh TP, Kumar RP. Human group III PLA2 as a drug target: structural analysis and inhibitor binding studies. Int J Biol Macromol. 2010;47(4):496–501. doi: 10.1016/j.ijbiomac.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26(5):680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Park D, Kim BC, Cho SW, Kim YT, Park YJ, et al. Predicting the interactome of Xanthomonas oryzae pathovar oryzae for target selection and DB service. BMC Bioinform. 2008;9:41. doi: 10.1186/1471-2105-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King LA, Nogareda F, Weill FX, Mariani-Kurkdjian P, Loukiadis E, Gault G, et al. Outbreak of Shiga toxin-producing Escherichia coli O104:H4 associated with organic fenugreek sprouts, France, June 2011. Clin Infect Dis. 2012;54(11):1588–1594. doi: 10.1093/cid/cis255. [DOI] [PubMed] [Google Scholar]
- Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39(Database issue):D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Nanduri B. HPIDB—a unified resource for host–pathogen interactions. BMC Bioinform. 2010;11(Suppl 6):S16. doi: 10.1186/1471-2105-11-S6-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Parvathi A, Hernandez RL, Cadle KM, Varela MF. Identification of a novel UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) from Vibrio fischeri that confers high fosfomycin resistance in Escherichia coli. Arch Microbiol. 2009;191(5):425–429. doi: 10.1007/s00203-009-0468-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavecchia A, Di Giovanni C. Virtual screening strategies in drug discovery: a critical review. Curr Med Chem. 2013;20(23):2839–2860. doi: 10.2174/09298673113209990001. [DOI] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1–3):3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Luo Q, Pagel P, Vilne B, Frishman D. DIMA 3.0: Domain Interaction Map. Nucleic Acids Res. 2011;39(Database issue):D724–D729. doi: 10.1093/nar/gkq1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356(6364):83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37(Database issue):D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michino H, Araki K, Minami S, Takaya S, Sakai N, Miyazaki M, et al. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am J Epidemiol. 1999;150(8):787–796. doi: 10.1093/oxfordjournals.aje.a010082. [DOI] [PubMed] [Google Scholar]
- Mondal SI, Khadka B, Akter A, Roy PK, Sultana R. Computer based screening for novel inhibitors against Vibrio cholerae using NCI diversity set-II: an alternative approach by targeting transcriptional activator ToxT. Interdiscip Sci Comput Life Sci. 2014;6(2):108–117. doi: 10.1007/s12539-012-0046-8. [DOI] [PubMed] [Google Scholar]
- Mondal SI, Ferdous S, Jewel NA, Akter A, Mahmud Z, Islam MM, et al. Identification of potential drug targets by subtractive genome analysis of Escherichia coli O157:H7: an in silico approach. Adv Appl Bioinform Chem AABC. 2015;8:49–63. doi: 10.2147/AABC.S88522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora A, Herrrera A, Lopez C, Dahbi G, Mamani R, Pita JM, et al. Characteristics of the Shiga-toxin-producing enteroaggregative Escherichia coli O104:H4 German outbreak strain and of STEC strains isolated in Spain. Int Microbiol Off J Span Soc Microbiol. 2011;14(3):121–141. doi: 10.2436/20.1501.01.142. [DOI] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35(Web Server issue):W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniesa M, Hammerl JA, Hertwig S, Appel B, Brussow H. Shiga toxin-producing Escherichia coli O104:H4: a new challenge for microbiology. Appl Environ Microbiol. 2012;78(12):4065–4073. doi: 10.1128/AEM.00217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Cheminformatics. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri WA, Jr, Miller M, Binder HJ, Levine MM, Dillingham R, Guerrant RL. Enteric infections, diarrhea, and their impact on function and development. J Clin Invest. 2008;118(4):1277–1290. doi: 10.1172/JCI34005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontius J, Richelle J, Wodak SJ. Deviations from standard atomic volumes as a quality measure for protein crystal structures. J Mol Biol. 1996;264(1):121–136. doi: 10.1006/jmbi.1996.0628. [DOI] [PubMed] [Google Scholar]
- Rajender PS, Vasavi M, Vuruputuri U. Identification of novel selective antagonists for cyclin C by homology modeling and virtual screening. Int J Biol Macromol. 2011;48(2):292–300. doi: 10.1016/j.ijbiomac.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Rao CV, Kirby JR, Arkin AP. Design and diversity in bacterial chemotaxis: a comparative study in Escherichia coli and Bacillus subtilis. PLoS Biol. 2004;2(2):E49. doi: 10.1371/journal.pbio.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JP, Levitt M, Chopra G. KoBaMIN: a knowledge-based minimization web server for protein structure refinement. Nucleic Acids Res. 2012;40(Web Server issue):W323–W328. doi: 10.1093/nar/gks376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar KR, Sakharkar MK, Chow VT. A novel genomics approach for the identification of drug targets in pathogens, with special reference to Pseudomonas aeruginosa. In silico Biol. 2004;4(3):355–360. [PubMed] [Google Scholar]
- Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32(Database issue):D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samland AK, Amrhein N, Macheroux P. Lysine 22 in UDP-N-acetylglucosamine enolpyruvyl transferase from Enterobacter cloacae is crucial for enzymatic activity and the formation of covalent adducts with the substrate phosphoenolpyruvate and the antibiotic fosfomycin. Biochemistry. 1999;38(40):13162–13169. doi: 10.1021/bi991041e. [DOI] [PubMed] [Google Scholar]
- Sliwoski G, Kothiwale S, Meiler J, Lowe EW., Jr Computational methods in drug discovery. Pharmacol Rev. 2014;66(1):334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CM, Fischer K, Abubucker S, Wang Z, Martin J, Jiang D, et al. Targeting protein–protein interactions for parasite control. PLoS One. 2011;6(4):e18381. doi: 10.1371/journal.pone.0018381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JR, Sharp JC, Dev VJ. Infection caused by Escherichia coli O157:H7 in Alberta, Canada, and in Scotland: a five-year review, 1987–1991. Clin Infect Dis. 1994;19(5):834–843. doi: 10.1093/clinids/19.5.834. [DOI] [PubMed] [Google Scholar]
- Wu S, Zhu Z, Fu L, Niu B, Li W. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genom. 2011;12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26(13):1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanzoni A, Montecchi-Palazzi L, Quondam M, Ausiello G, Helmer-Citterich M, Cesareni G. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513(1):135–140. doi: 10.1016/S0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- Zhang R, Lin Y. DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res. 2009;37(Database issue):D455–D458. doi: 10.1093/nar/gkn858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Ou HY, Zhang CT. DEG: a database of essential genes. Nucleic Acids Res. 2004;32(Database issue):D271–D272. doi: 10.1093/nar/gkh024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


