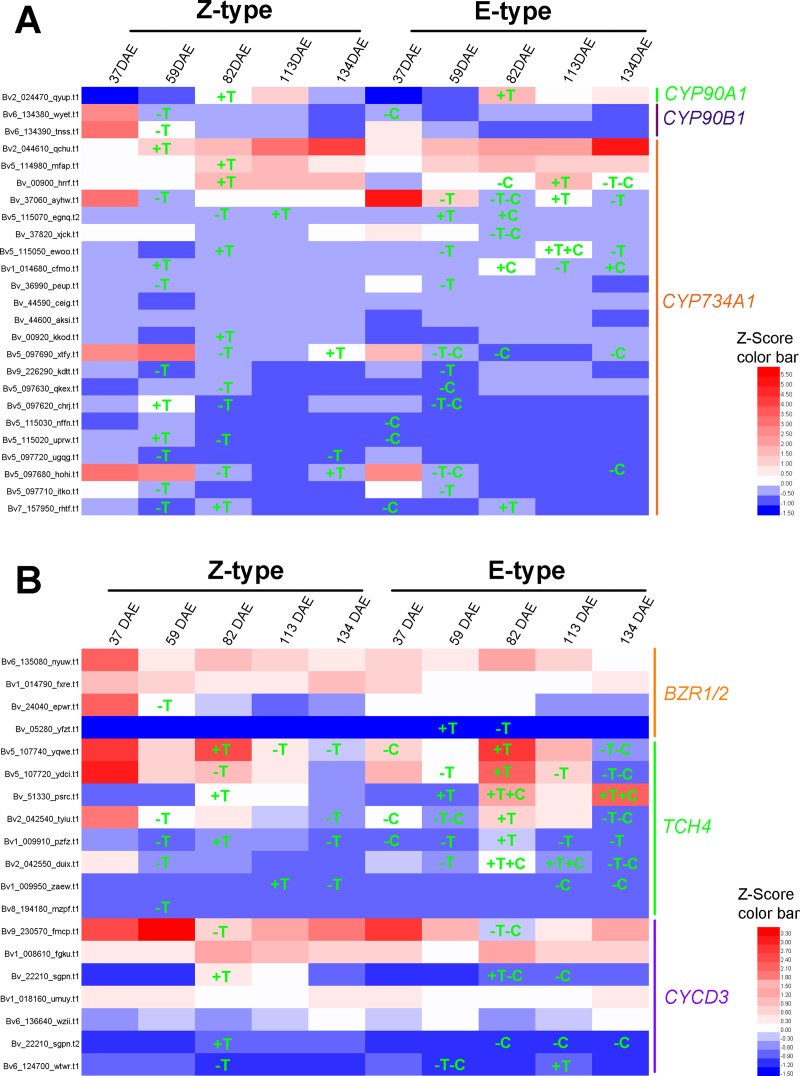
Fig 6. Expression pattern of genes involved in the brassinosteroid signaling.
(A) Orthologues of genes encoding the key enzymes for the biosynthesis and metabolism of brassinosteroid. (B) Orthologues of genes encoding the components in the signal transduction pathways of brassinosteroid. Z score transformation [26] was calculated by subtracting the mean FPKM of all the “main Beta vulgaris orthologs” in all of the ten samples, and dividing that result by the standard deviation based on all the “main Beta vulgaris orthologs” in all of the ten samples for each gene. The “main Beta vulgaris orthologs” included the DEGs or the genes with FPKM ≥1 in any library. Gene IDs were marked on the left. +, up-regulated; -, down-regulated; T, up- and down-regulated compared the previous growth stage; C, E-type compared with Z-type.