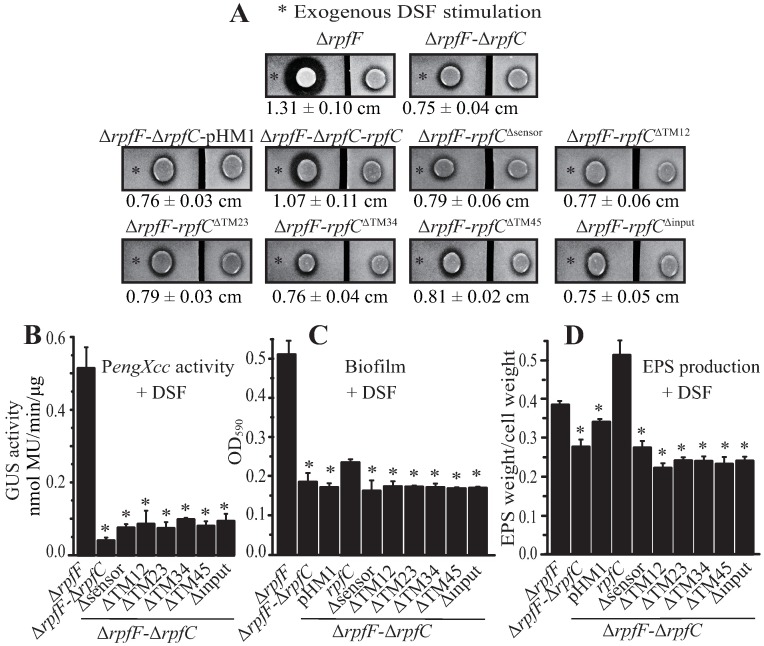
Fig 2. The input region of RpfC is essential in DSF sensing.
(A) Putative sensor and transmembrane helices contribute to DSF detection. In each panel, duplicate bacterial colonies were inoculated on the left and right and were separated by cutting the NYG plate into two parts. A total of 3.5 μL of DSF (30 μM, indicated by an asterisk) were spotted near the colony on the left. The ability to produce extracellular proteases was observed after 36 h of incubation. Average diameters (cm) of the protein degradation zones of the left colony with standard deviations (n = 4) are listed below each panel. (B, C, and D) Mutation of the input region of rpfC compromised the ability of the bacteria to detect DSF and modulate the expression of engXcc (B), form a biofilm (C), or produce EPS (D). In (B), each bacterial strain contains a recombinant pHM2 vector with a PengXcc-GUS transcriptional fusion. In (B, C, and D), 10 μM exogenous DSF was added to bacterial cultures. GUS activities, EPS production, and biofilm development were measured after 9 h, 75 h and 12 h, respectively. Black bars represent standard deviation (n = 3). * = statistically significant (P ≤ 0.05) compared with the ΔrpfF strain, measured by the Student’s t-test.