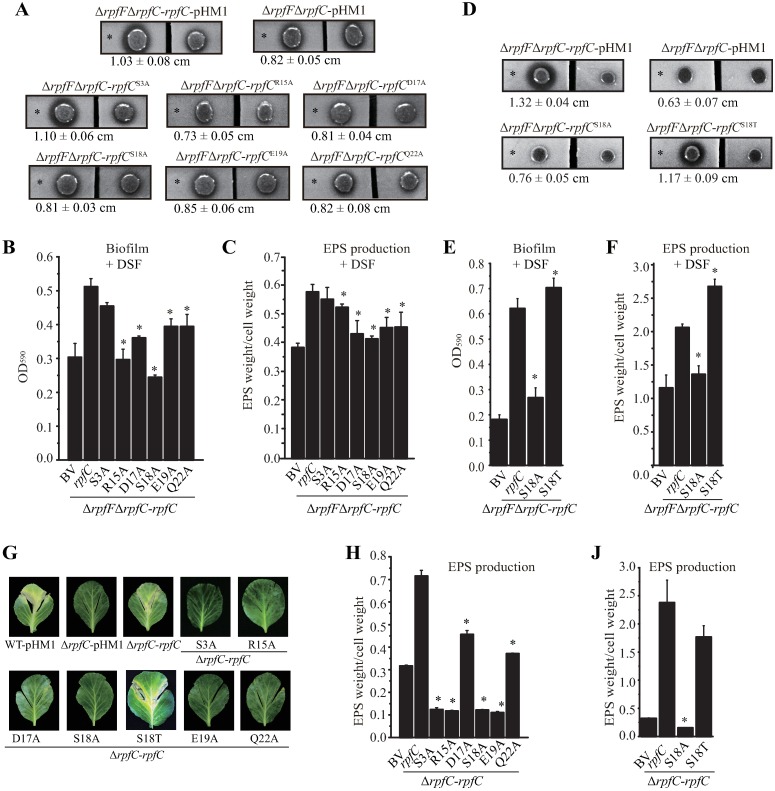
Fig 5. Impact of substitution of essential amino acids in RpfC sensor on DSF perception and bacterial virulence.
(A and D). Amino acid substitution affects extracellular protease activity of bacteria. In each panel, bacterial colonies on the left and right are the same but were separated by cutting the NYG plate into two parts. A total of 3.5 μL of DSF (30 μM, indicated by an asterisk) were spotted near the left colony. Recovery of the ability to produce extracellular proteases was observed after 36 h of incubation. Average diameters (cm) of the protein degradation zones with standard deviations (n = 4) are listed below each panel. (B and E). Amino acid substitution affects biofilm formation of bacteria. Quantification of biofilm was performed by crystal violet staining methods. DSF was added to each bacterial culture at 10 μM prior to biofilm quantification. * indicates significant different to that of the control (ΔrpfFΔrpfC-rpfC, n = 4), calculated by a Student’s t-test (P ≤ 0.05). (C, F, H and J). Amino acid substitution affects exopolysaccharides (EPS) formation of bacteria. Bacterial strains were cultured in NYG medium with 10 μM DSF for 72 hours (C, and H) or 96 hours (F and J) before EPS quantification. * indicates significant different to that of the control (ΔrpfFΔrpfC-rpfC, n = 4), calculated by a Student’s t-test (P ≤ 0.05). (G) Bacterial virulence against host plant B. oleraceae cv. Jingfeng No. 1. Eight-week old plant was inoculated with bacterial cultures by sterile scissors. The levels of bacterial virulence were estimated 10 days after inoculation. A semi-quantification of virulence scale was shown in S3B and S3C Fig BV: ΔrpfFΔrpfC strain containing a blank pHM1 vector.