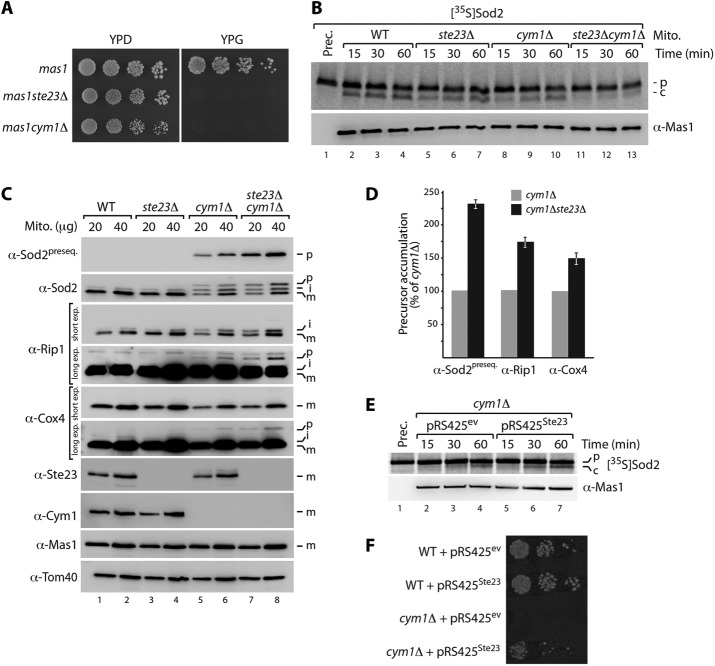
FIGURE 4:
Peptide clearance by Ste23 and Cym1 is required for functional presequence processing machinery. (A) Synthetic lethality of mas1ste23Δ and mas1cym1Δ double mutants. Strains were grown under fermentative (YPD) or respiratory (YPG) conditions at 23°C. (B) Presequence-processing activity in soluble mitochondrial extracts from indicated yeast strains was analyzed by incubation with radiolabeled Sod2 precursor, followed by SDS–PAGE and autoradiography. c, cleaved Sod2 protein; p, precursor. (C) Immunoblot analysis of mitochondria isolated from WT, ste23Δ, cym1Δ, and ste23Δcym1Δ yeast cells. i, processing intermediate; m, processed, mature protein. (D) Levels of nonprocessed preproteins (p) analyzed in C were quantified. Quantifications represent mean ± SEM (n = 3). (E) Overexpression of Ste23 in cym1Δ cells stimulates presequence processing of radiolabelled Sod2 precursor in soluble mitochondrial extracts. c, cleaved Sod2 protein; ev, empty vector; p, precursor. (F) Ste23 rescues loss of Cym1 function in vivo. Overexpression of Ste23 from pRS425 plasmid restores viability of cym1Δ cells at 37°C.