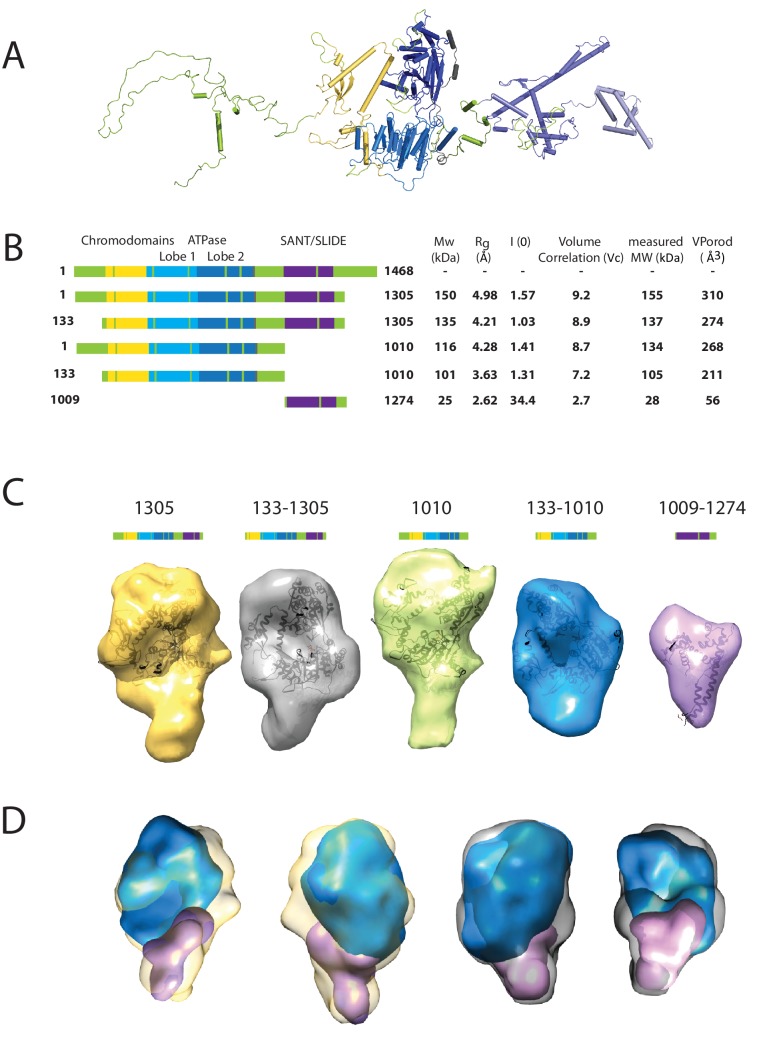
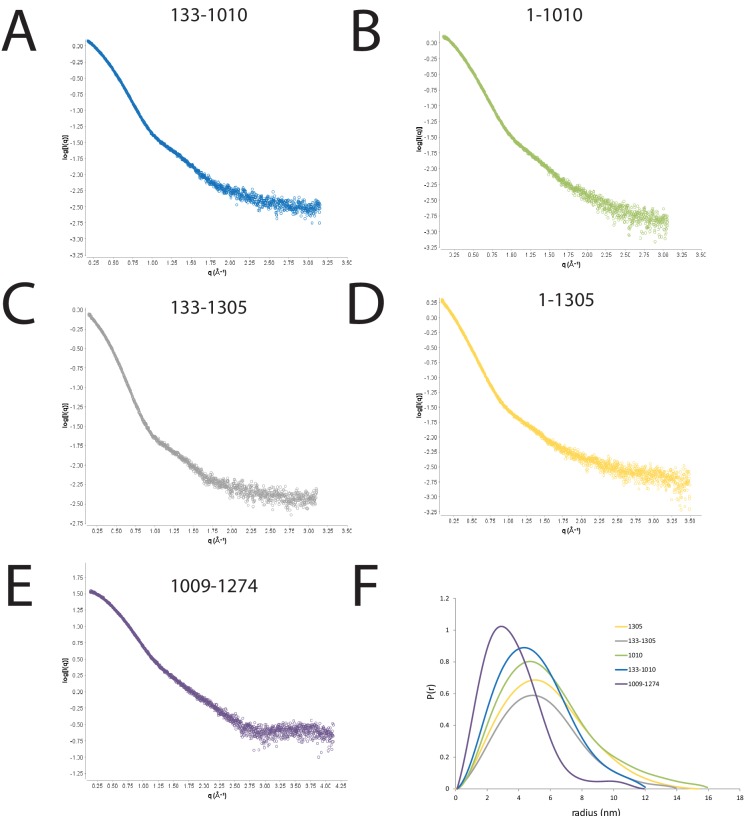
Figure 1. Characterising the solution structure of Chd1 by small angle X-ray scattering (SAXS).
(A) The overall structure of S. cerevisiae Chd1 molecule. Previously characterised structural features including the chromoATPase (3MWY) (Hauk et al., 2010) with chromodomains coloured yellow, ATPase lobe 1 - marine, ATPase lobe 2 – blue. The DNA binding domain (2XB0) (Ryan et al., 2011) is coloured deep blue with the NMR structure of the C-terminal extension (2N39) (Mohanty et al., 2016) in pale blue. The unresolved structural elements are coloured pale green and a cartoon representation of their predicted secondary structure provided to give an idea of scale. (B) On the left Illustration of various Chd1 truncations. The known domains within the truncation are labelled and coloured. On the right, the data (hydrodynamic radius (Rg), extrapolated zero intensity (I(0)), molecular weight (MW)) obtained from the SAXS analysis of the respective construct protein. (C) Ab-initio bead models generated from the one-dimensional scattering curves using GASBOR for different constructs. Known Chd1 crystal structures are docked into the respective volume. (D) The volumes for 133–1010 (blue) and 1009–1274 (pink) are fitted into 1305 (yellow) and 133–1305 (grey) volume maps using SASREF.
Figure 1—figure supplement 1. Purification of fragments of Chd1 protein.