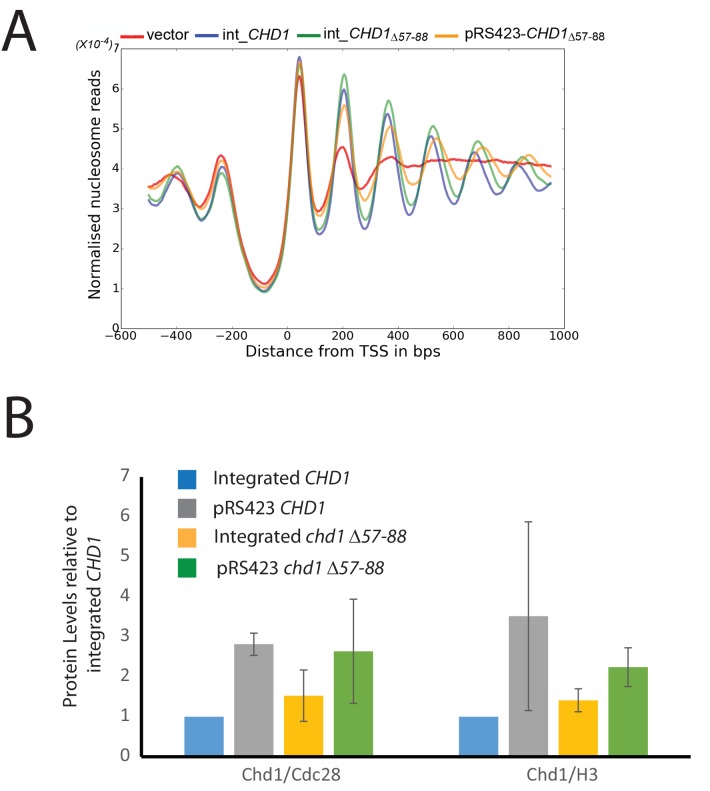
(
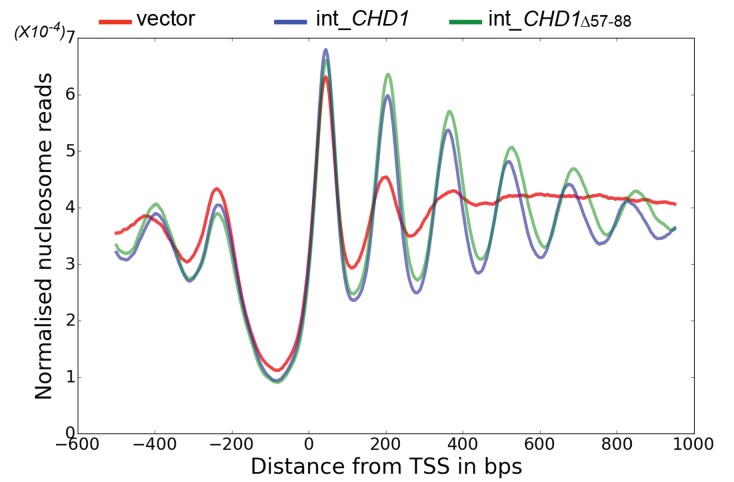
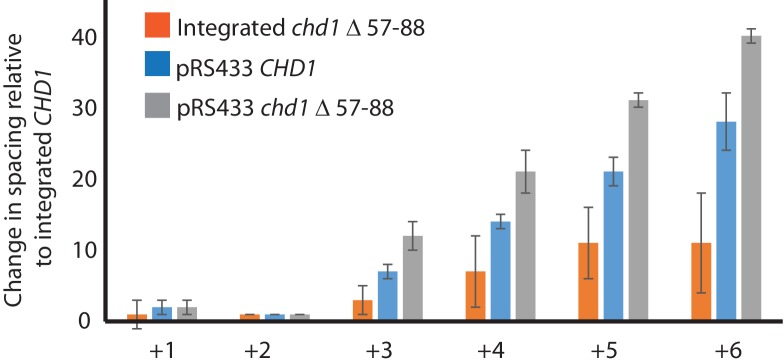
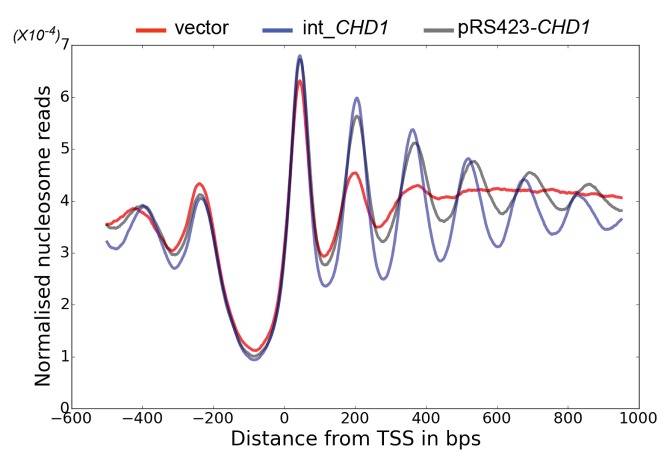
A) Changes in nucleosome spacing in coding regions were measured in strains expressing
chd1△57–88 re-integrated in single copy at the
CHD1 locus (int_CHD1△57–88, green) or at higher copy number on pRS423 (pRS423_CHD1△57–88, orange). Vector only (red) and
CHD1 wt reintroduced integrated onto the chromosome in single copy (int-
CHD1, blue) from
Figure 5 are also shown for comparison. TSS-aligned nucleosome occupancy profiles show increased spacing in strains with
chd1△57–88 compared to wild-type
CHD1 (int_
CHD1). Replacement on a high copy plasmid results in two- to fourfold increased Chd1 expression. (
B) Quantitative measurement of protein levels in strains expressing
CHD1wt or
chd1△57–88 at low (int) or high (pRS423) copy number. For all strains described in A, Chd1 was N-terminally flag tagged and anti-FLAG Western blotting used to plot protein levels relative to int-
CHD1wt. Quantitation is plotted following normalisation against total histone H3 and Cdc28 as indicated. Error bars indicate standard error from 3–4 measurements.