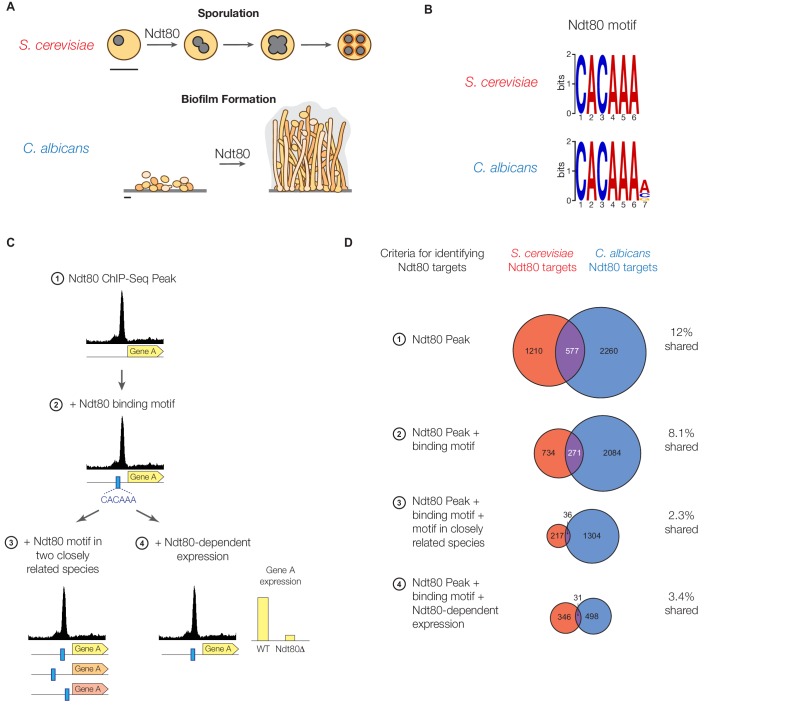
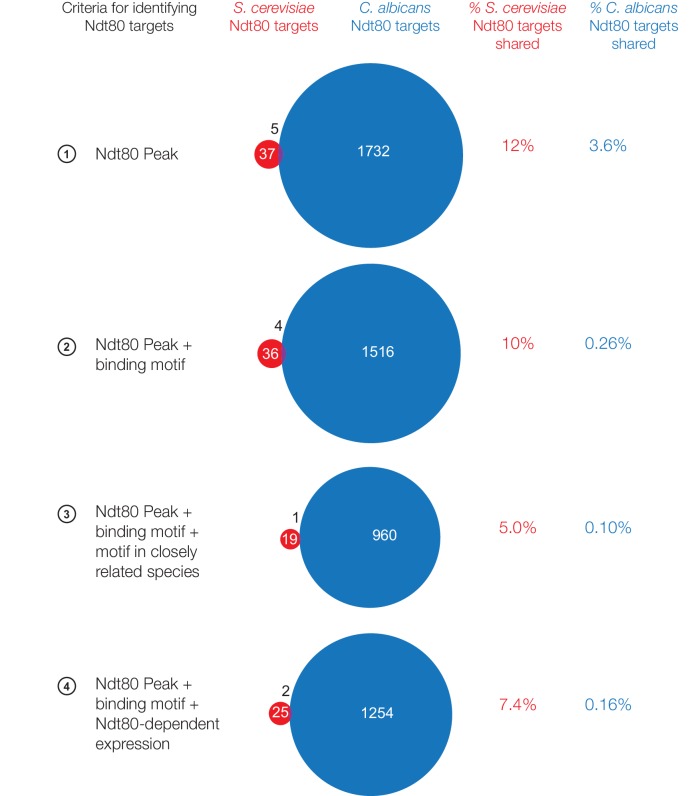
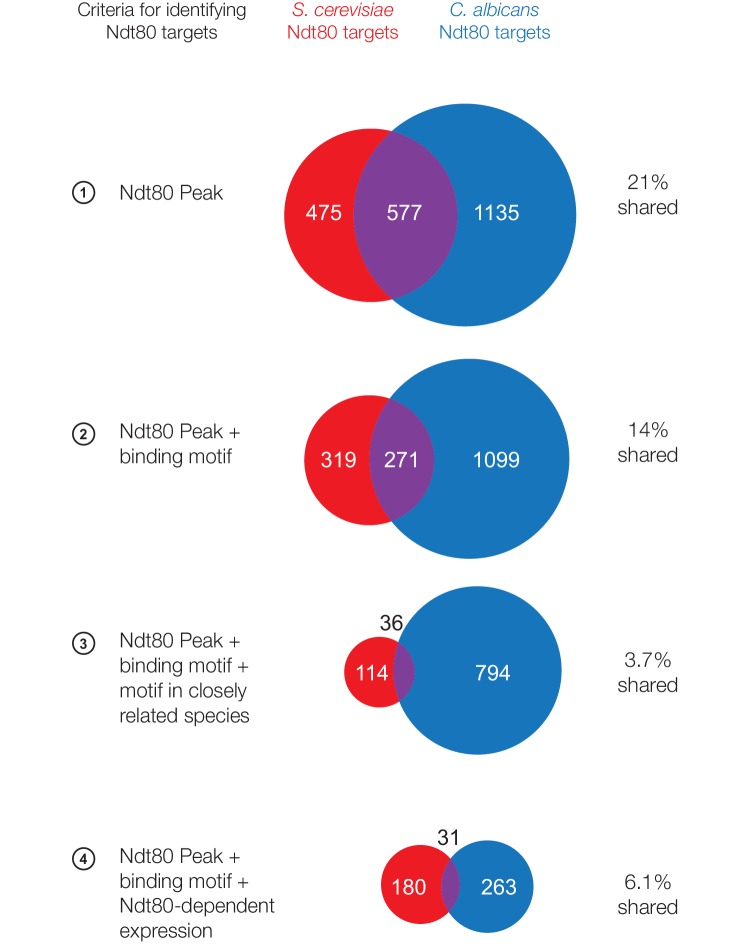
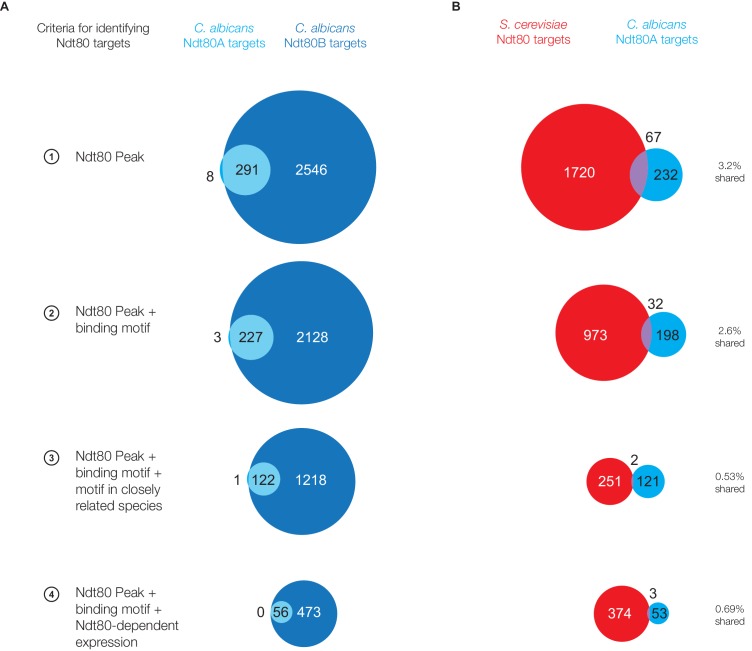
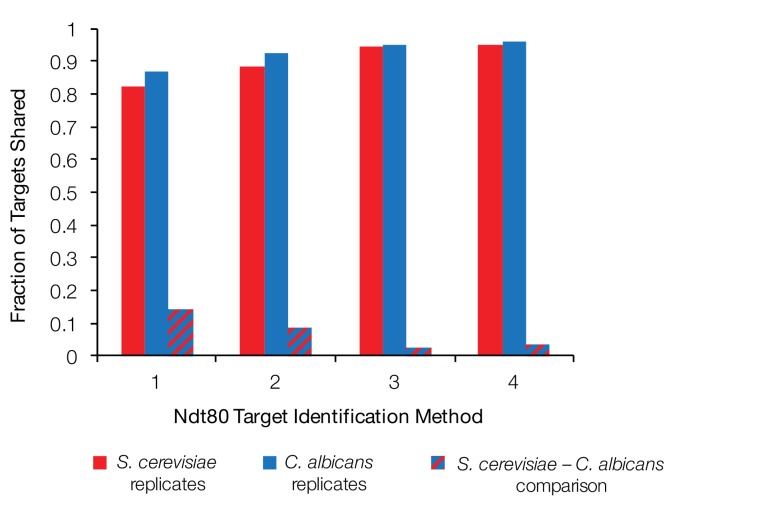
Figure 1. Ndt80 target genes differ between S. cerevisiae and C. albicans.
(A) Diagram of sporulation in S. cerevisiae and biofilm formation in C. albicans. Scale bars represent 5 µm. (B) The cis-regulatory motif most highly enriched at locations of Ndt80 ChIP binding in S. cerevisiae and C. albicans. Motifs were generated independently for each species using DREME. The Ndt80 motifs determined de novo in this study closely match those identified previously for Ndt80 (Chu and Herskowitz, 1998; Jolly et al., 2005; Nobile et al., 2012). (C) Diagram of the four criteria used to identify Ndt80 regulatory targets. Criteria 1: significant ChIP-Seq enrichment in the intergenic region upstream of a gene relative to untagged control experiments. Criteria 2: ChIP-Seq enrichment and the presence of an Ndt80 motif in the intergenic region. Criteria 3: ChIP-Seq enrichment with the Ndt80 motif present in the intergenic region and also in orthologous intergenic regions of two very closely related species, suggesting the motif has been maintained by selection. Criteria 4: ChIP-Seq enrichment with the Ndt80 motif present in the intergenic region and Ndt80-dependent expression of the nearby gene, indicating that expression of the gene is under Ndt80 control. (D) Overlap in targets of S. cerevisiae Ndt80 (red) and C. albicans Ndt80B (blue), using the four different criteria from (C) to identify targets, when Ndt80 is highly expressed in each species (Materials and methods). Venn diagrams are roughly area-proportional (Hulsen et al., 2008).