Summary
Chromatin and metabolic states both influence lifespan, but how they interact in lifespan regulation is largely unknown. The COMPASS chromatin complex, which trimethylates lysine 4 on histone H3 (H3K4me3), regulates lifespan in C. elegans. However, the mechanism by which H3K4me3 modifiers impact longevity, and whether it involves metabolic changes, remain unclear. Here we find that H3K4me3-methyltransferase deficiency, which extends lifespan, promotes fat accumulation with a specific enrichment of mono-unsaturated fatty acids (MUFAs). This fat metabolism switch in H3K4me3-methyltransferase deficient animals is mediated at least in part by downregulation of germline targets, including S6 kinase, and by activation of an intestinal transcriptional network that upregulates delta-9 fatty acid desaturases. Interestingly, MUFA accumulation is necessary for the lifespan extension of H3K4me3-methyltransferase deficient worms, and dietary MUFAs are sufficient to extend lifespan. Given the conservation of lipid metabolism, dietary or endogenous MUFAs could extend lifespan and healthspan in other species, including mammals.
Introduction
Chromatin state, which encompasses post-translational modification of histone proteins, integrates environmental signals to influence gene expression and downstream cellular processes. Enzymes that deposit or remove histone modifications regulate lifespan in several species1,2. For example, methyltransferase/demethylase complexes that modify H3K4me3, H3K27me3, and H3K36me3 modulate lifespan in yeast, worms, and flies1,2. Several chromatin-modifying enzymes require intracellular metabolites as co-factors (e.g. NAD+, α-ketoglutarate, or S-adenosyl methionine), rendering their activity exquisitely sensitive to metabolic states1,2. A subset of these chromatin-modifying enzymes, such as deacetylases, in turn influence metabolism1,2. However, the impact of methyltransferases and demethylases on metabolism remains unexplored.
Fat metabolism plays an important role in many physiological and pathological processes. It influences long-term energy storage, inter- and intra-cellular signaling, and membrane homeostasis. In humans, excessive fat storage in the form of triglycerides is associated with diseases such as atherosclerosis and type 2 diabetes3. On the other hand, emerging studies in both invertebrates and mammals suggest that specific alterations in fat profiles, and even elevated fat storage, can be associated with longevity4. Whether fat metabolism links chromatin modifiers to lifespan extension is unknown.
H3K4me3 modifiers impact fat metabolism
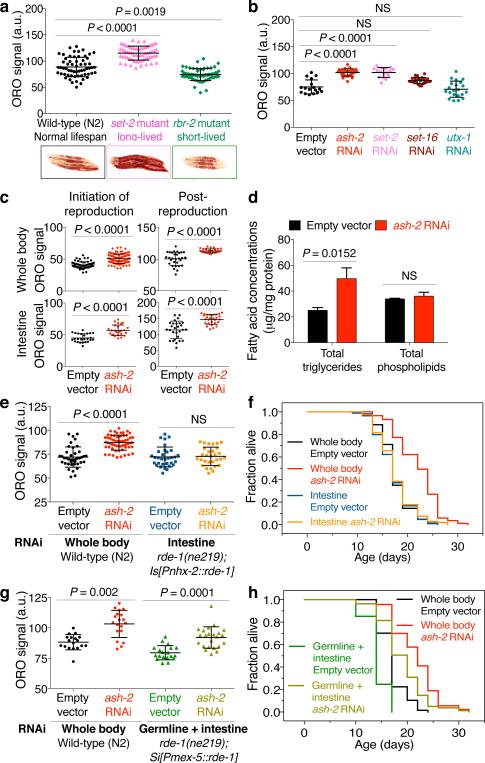
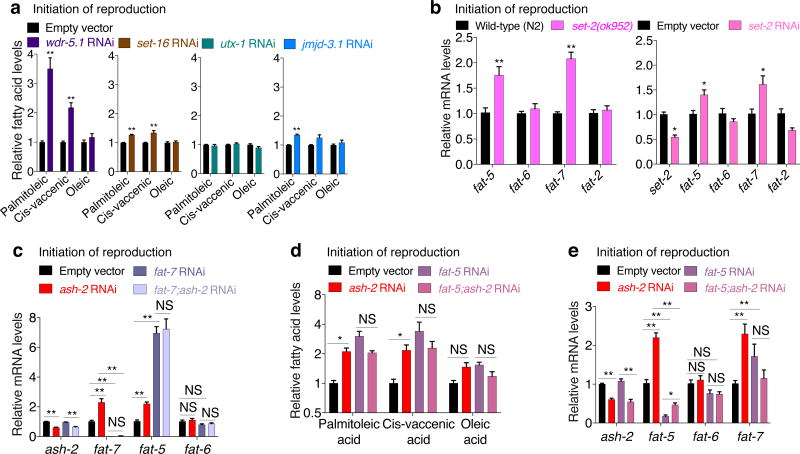
The COMPASS chromatin complex, which catalyzes trimethylation of lysine 4 on histone H3 (H3K4me3)5, regulates lifespan in C. elegans6. The H3K4me3 global landscape is remodeled during aging and in cellular senescence in mammals1,2. However, the mechanism by which H3K4me3 modifiers impact longevity remains unclear. To determine whether the COMPASS H3K4me3 methyltransferase complex influences fat metabolism, we used Oil-Red-O (ORO) staining. In fixed tissues, ORO stains neutral lipids such as triglycerides (TAGs)7. Interestingly, worms with deficiency in the COMPASS H3K4me3 methyltransferase SET-2/SET1, which are long-lived6 (Supplementary Table 1), exhibit increased ORO staining (Fig. 1a). Conversely, worms deficient for the H3K4me3 demethylase RBR-2/JARID1, which are slightly short-lived6 (Supplementary Table 1), display decreased ORO staining (Fig. 1a). RNAi knockdown of ash-2/ASH2L and wdr-5.1/WDR5, shared components between the COMPASS and Trithorax-related H3K4me3 complexes5, and of set-2, a specific component of the COMPASS H3K4me3 complex5, also extended lifespan6 (Supplementary Table 1) and increased ORO staining (Fig. 1b, Extended Data Fig. 1a). In contrast, knockdown of set-16/MLL and utx-1/UTX, specific components of the Trithorax-related H3K4me3 complex5, did not significantly affect ORO levels (Fig. 1b). Thus, the COMPASS H3K4me3 methyltransferase complex specifically influences fat levels.
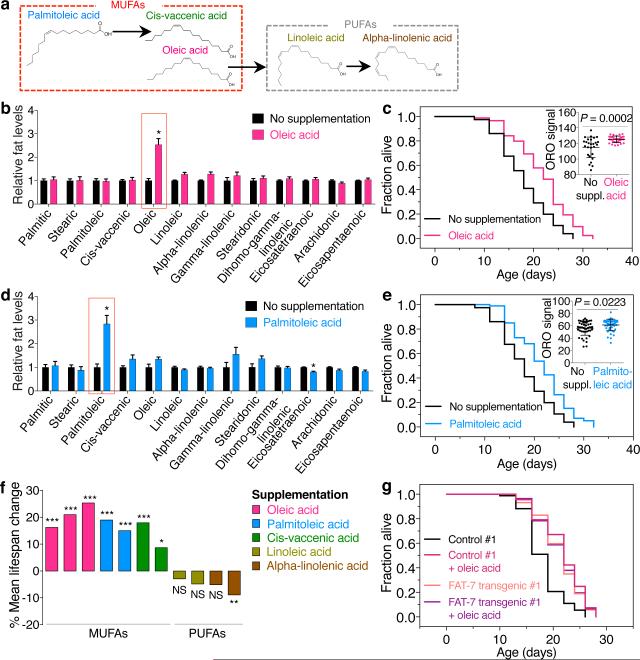
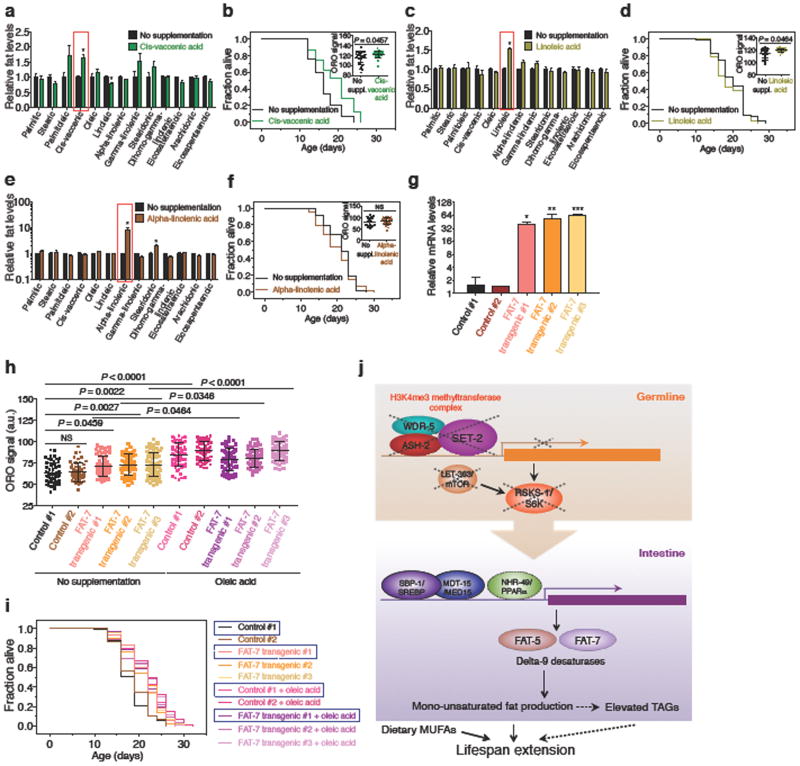
Figure 1. H3K4me3-methyltransferase deficiency in the germline leads to fat accumulation in the intestine.
a, ORO staining images and quantification. Mean ± s.d., n ≥ 55 worms per condition. b, As in a, n ≥ 16 worms per condition. c, As in a, n ≥ 29 worms (upper panels) or n ≥ 25 intestinal cells (lower panels) per condition. d, Gas chromatography-mass spectrometry (GC-MS) quantification. Mean ± s.e.m of 2 independent experiments, each with 3 biological replicates. e, As in a, n ≥ 26 worms per condition. f, ash-2 RNAi extends lifespan in wild-type (P < 0.0001, log-rank) but not in rde-1(ne219); Is[Pnhx-2::rde-1] worms. g, As in a, n ≥ 18 worms per condition. h, ash-2 RNAi extends lifespan in both wild-type and rde-1(ne219); Si[Pmex-5::rde-1] worms (P < 0.0001, log-rank). b, c, e-h, Representative of 2 experiments. P values: a, b, e, g, Kruskal-Wallis with Dunn's correction; c, d, Two-tailed Mann-Whitney.
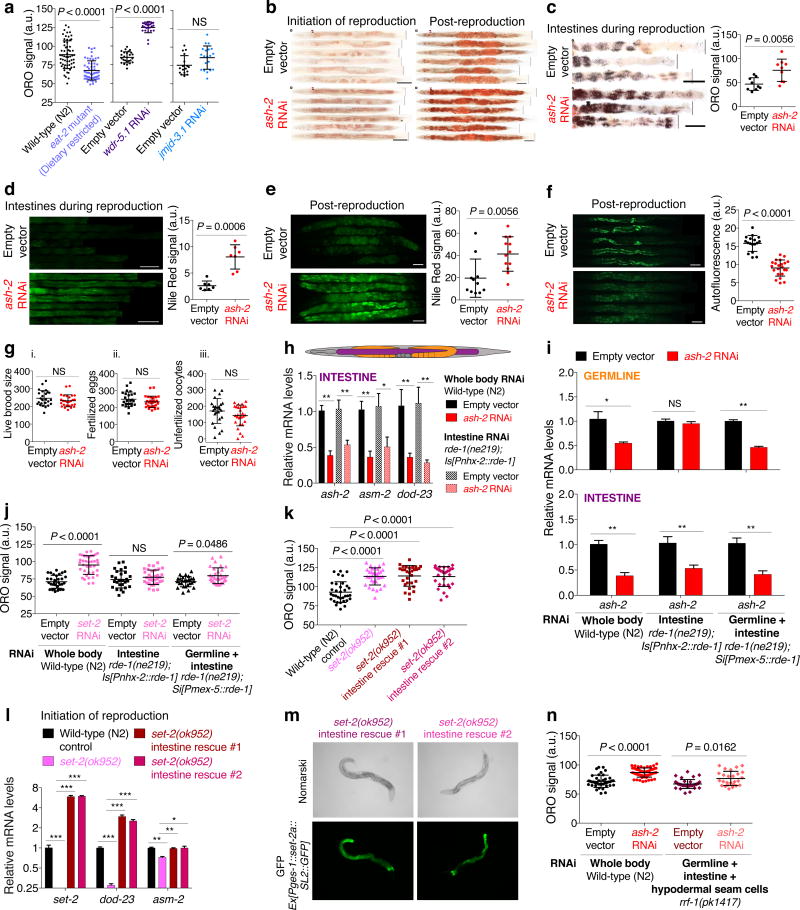
Fat accumulated most prominently in the intestine in ash-2 knockdown animals (Fig. 1c), and this was observed at the initiation of reproduction, during reproduction, and post-reproduction (Fig. 1c, Extended Data Fig. 1b, c). The intestinal fat accumulation in ash-2 knockdown worms was also observed using Nile Red, which stains neutral lipids in fixed tissues8 (Extended Data Fig. 1d-f). Furthermore, gas chromatography coupled to mass spectrometry (GC-MS) showed that ash-2 knockdown worms have significantly higher levels of TAGs, but similar levels of phospholipids (PLs), compared to controls (Fig. 1d). Fat accumulation in ash-2 knockdown worms is unlikely to be caused by a shift in fat resources resulting from reduced fertility, because ash-2 knockdown animals have a similar number of progeny as wild-type animals6 (Extended Data Fig. 1g). Thus, H3K4me3-methyltransferase deficient worms, which are long-lived and fertile, accumulate intestinal fat in the form of TAGs.
H3K4me3 modifiers act mostly in the germline to extend lifespan6, yet fat accumulation occurs in the intestine (Fig. 1c). To determine whether H3K4me3 modifiers act in the germline or intestine to regulate fat accumulation, we performed tissue-selective RNAi knockdown and transgenic rescue experiments. Knockdown of ash-2 in the intestine (rde-1(ne219); Is[Pnhx-2::rde-1]9) did not promote fat accumulation or lifespan extension (Fig. 1e, f, Extended Data Fig. 1h, i). Likewise, set-2 knockdown in the intestine did not increase fat accumulation (Extended Data Fig. 1j), and overexpression of set-2 in the intestine of set-2(ok952) mutants did not abrogate the elevated fat levels of set-2(ok952) mutants (Extended Data Fig. 1k-m). These results suggest that H3K4me3 modifiers act mostly outside of the intestine to influence fat accumulation. As strains that allow germline-specific RNAi knockdown do not currently exist in C. elegans, we used the rde-1(ne219);Si[Pmex-5::rde-1] strain, which restricts RNAi to germline and intestine10. Interestingly, ash-2 knockdown in germline and intestine promoted fat accumulation and extended lifespan (Fig. 1g, h, Extended Data Fig. 1i). Likewise, set-2 knockdown in germline and intestine also increased fat levels (Extended Data Fig. 1j). Similarly, ash-2 knockdown in rrf-1(pk1417) mutants, in which RNAi is restricted to the germline, intestine, and some hypodermal seam cells11, also increased fat levels and extended lifespan (Extended Data Fig. 1n, Supplementary Table 1). Thus, the COMPASS H3K4me3 methyltransferase complex acts mostly in the germline to regulate intestinal fat accumulation and lifespan, implying a germline-to-intestine communication.
A switch to MUFA metabolism
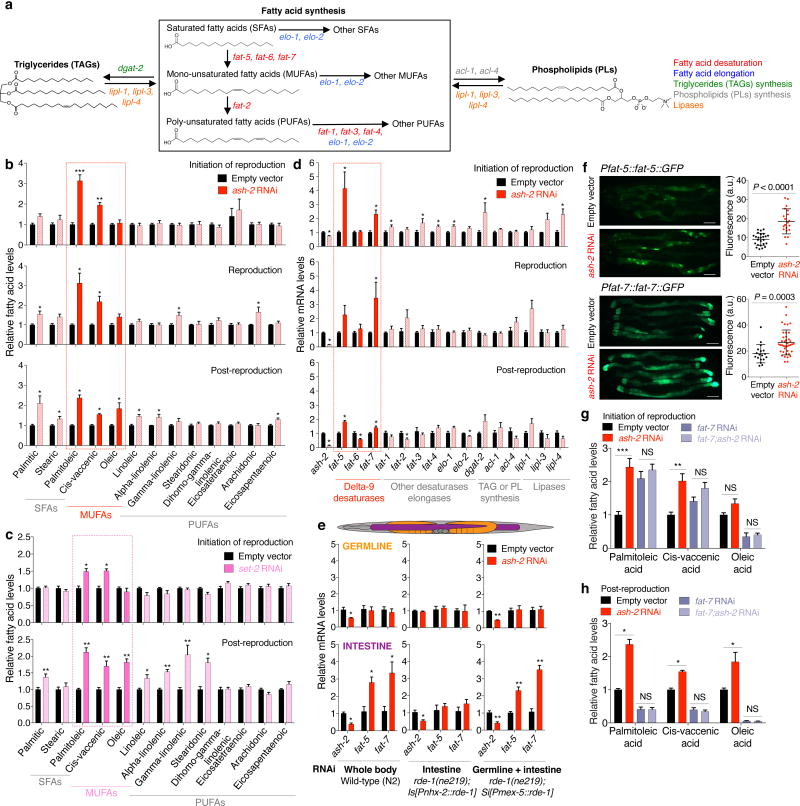
Like mammals, worms have different categories of fatty acids: saturated fatty acids (SFAs), mono-unsaturated fatty acids (MUFAs), and poly-unsaturated fatty acids (PUFAs) (Fig. 2a). To identify specific lipid species associated with longevity in H3K4me3-methyltransferase deficient worms, we profiled long-chain fatty acids by GC-MS. Interestingly, at all ages tested, ash-2 knockdown increased MUFAs, while SFAs and PUFAs remained mostly unchanged (Fig. 2b). The MUFAs palmitoleic acid and cis-vaccenic acid were significantly elevated at all ages, whereas the MUFA oleic acid was raised post-reproduction in ash-2 knockdown worms and in worms deficient for set-2, a specific component of the COMPASS H3K4me3 complex (Fig. 2b, c). In contrast, MUFAs remained largely unaffected in worms deficient for set-16 and utx-1, specific components of the Trithorax-related H3K4me3 complex (Extended Data Fig. 2a). Thus, the COMPASS H3K4me3 methyltransferase complex specifically impacts MUFA metabolism.
Figure 2. H3K4me3-methyltransferase deficiency results in elevated MUFAs and upregulation of delta-9 desaturases.
a, Fat metabolism pathways with genes used in RT-qPCR. b-c, GC-MS quantification of fatty acids. Mean ± s.e.m. of 2-3 independent experiments, each with 3 biological replicates. d, RT-qPCR. Mean ± s.e.m. of 2 independent experiments, each with 3 biological replicates. Solid red bars: delta-9 desaturases. e, RT-qPCR. Mean ± s.e.m. of 2 independent experiments, each with 2-3 biological replicates. f, Images and quantification of GFP fluorescence. Mean ± s.d., n ≥ 18 worms per condition. Representative of 2 (FAT-5) and 3 (FAT-7) experiments. Scale bar: 100 μm. g-h, GC-MS quantification of MUFAs. Mean ± s.e.m. of 3 (g) and 2 (h) independent experiments, each with 2-3 biological replicates. P values: Two-tailed Mann-Whitney (with Benjamini-Hochberg correction for > 2 conditions). * P < 0.05, ** P < 0.01, *** P < 0.001.
To determine whether this switch to MUFA accumulation arises from changes in fat synthesis, storage, or breakdown, we examined the expression of the specific enzymes that control these steps (Fig. 2a). ash-2 knockdown strongly increased the expression of delta-9 fatty acid desaturase genes fat-5 and fat-7 (Fig. 2d). FAT-5, a palmitoyl-CoA desaturase, preferentially converts palmitic acid into the MUFA palmitoleic acid, whereas FAT-7 and FAT-6, two stearoyl-CoA desaturases, convert stearic acid into the MUFA oleic acid12 (Fig. 2a). Expression of fat-5 and fat-7 was also upregulated in set-2 mutant or knockdown worms (Extended Data Fig. 2b). In contrast, the expression of fat-2, which catalyzes the first step in converting MUFAs to PUFAs13, and that of other fat metabolism enzymes (e.g. lipases), was not consistently affected by ash-2 knockdown (Fig. 2d). Expression of fat-5 and fat-7 was elevated in the intestine, but not in the germline, of ash-2 knockdown worms (Fig. 2e), and ash-2 acts mostly in the germline to promote this intestinal fat-5 and fat-7 elevation (Fig. 2e). Transgenic worms expressing green fluorescent protein (GFP) translational reporters for FAT-5 or FAT-7 also showed increased GFP fluorescence in the intestine upon ash-2 knockdown (Fig. 2f). Thus, germline H3K4me3-methyltransferase deficiency leads to upregulation of delta-9 desaturases in the intestine, suggesting that MUFA accumulation occurs at the level of synthesis.
We next examined the role of delta-9 desaturases in the MUFA accumulation of H3K4me3-methyltransferase deficient worms. Knockdown of fat-7 in ash-2 deficient worms specifically decreased oleic acid levels at the initiation of reproduction and reduced all three MUFAs (oleic, palmitoleic, and cis-vaccenic acid) post-reproduction (Fig. 2g, h, Extended Data Fig. 2c). In contrast, fat-5 knockdown in ash-2 deficient worms increased palmitoleic and cis-vaccenic acid (Extended Data Fig. 2d), likely due to compensatory upregulation of fat-7 (Extended Data Fig. 2e). These results suggest that the MUFA switch in H3K4me3-methyltransferase deficient worms is driven by FAT-7 upregulation in the intestine.
Germline targets of H3K4me3 modifiers
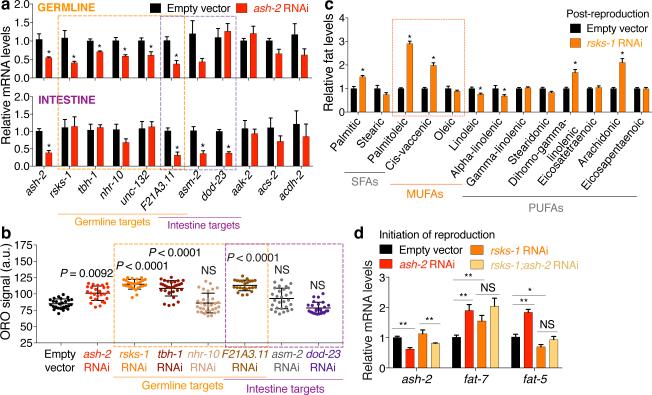
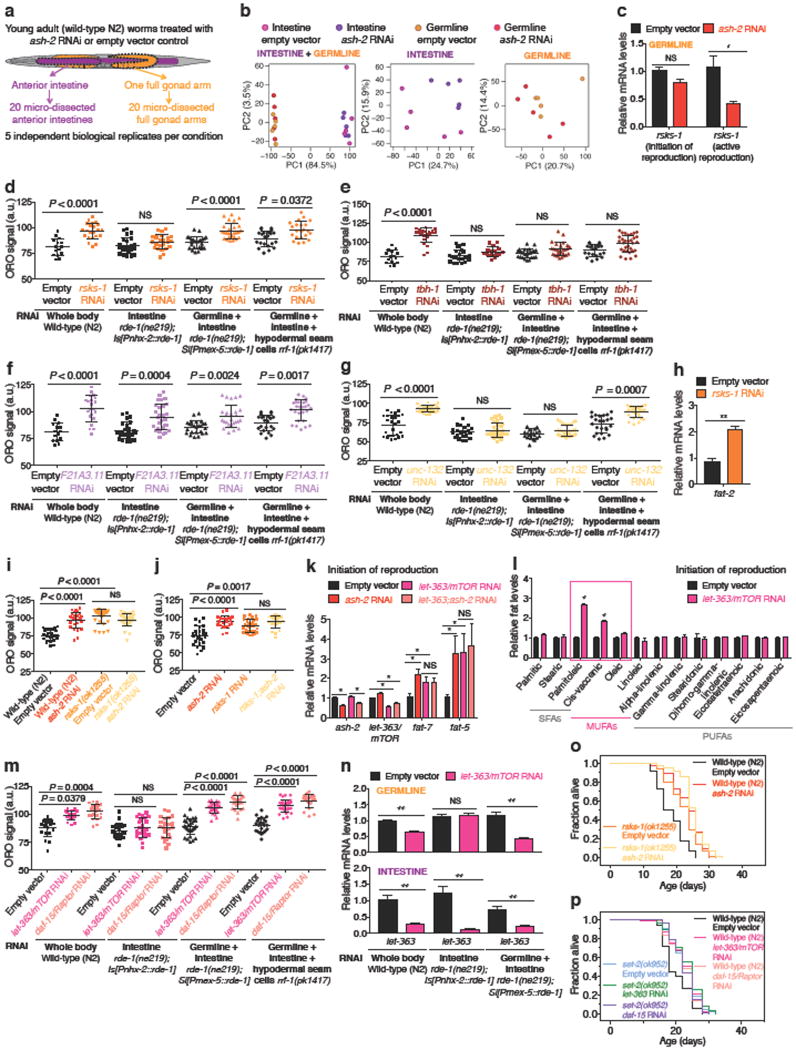
The H3K4me3 methyltransferase complex acts mostly in the germline to trigger intestinal upregulation of delta-9 desaturases. To better understand this germline-to-intestine communication, we identified germline targets of the H3K4me3 methyltransferase complex. We generated an RNA-seq dataset using micro-dissected germlines and intestines from ash-2 knockdown worms and analyzed this dataset (Extended Data Fig. 3a, b, Supplementary Table 2-5) as well as other large-scale datasets6,14,15. Because H3K4me3 methyltransferase complexes generally activate gene expression5, we selected targets that were downregulated in H3K4me3-methyltransferase deficient worms (Extended Data Table 1). Using RT-qPCR, we validated that ash-2 knockdown led to the significant downregulation of four candidates in the germline (rsks-1/S6 kinase, tbh-1, nhr-10, and unc-132), one candidate in both germline and intestine (F21A3.11), and two candidates in the intestine (asm-2 and dod-23) (Fig. 3a, Extended Data Fig. 3c).
Figure 3. H3K4me3-methyltransferase germline targets impact fat metabolism.
a, RT-qPCR. Mean ± s.e.m. of 2 independent experiments, each with 3 biological replicates. b, ORO quantification. Mean ± s.d., n ≥ 23 worms per condition. Representative of 2 experiments. c, GC-MS profiles of fatty acids. Mean ± s.e.m. of 2 independent experiments, each with 2-3 biological replicates. d, RT-qPCR. Mean ± s.e.m. of 2 independent experiments, each with 3 biological replicates. P values: b, Kruskal-Wallis with Dunn's correction; a, c, d, Two-tailed Mann-Whitney with Benjamini-Hochberg correction. * P < 0.05, ** P < 0.01.
We next assessed the functional involvement of these candidates in fat metabolism. Knockdown of rsks-1, tbh-1, F21A3.11, or unc-132 all led to increased ORO staining (Fig. 3b, Extended Data Fig. 3d-g). Among these four targets, rsks-1 was the only one that acted mostly in the germline to regulate fat accumulation (Extended Data Fig. 3d-g). RSKS-1/S6 kinase is a key conserved substrate of mTOR complex 116. RSKS-1 deficiency extends lifespan in C. elegans and mice17 and promotes fat accumulation in C. elegans18. We asked whether rsks-1 could mediate the fat metabolism switch of H3K4me3-methyltransferase deficient worms. rsks-1 knockdown or mutation led to increased fat accumulation, MUFA levels, and fat-7 expression (Fig. 3b-d, Extended Data 3h-j). Interestingly, ash-2 knockdown could not further increase fat accumulation and fat-7 expression in rsks-1 deficient worms (Fig. 3d, Extended Data 3i, j). Likewise, knockdown of the RSKS-1 upstream regulator, let-363/mTOR, also led to increased fat accumulation, MUFA levels, and fat-7 expression (Extended Data Fig. 3k-n), and ash-2 knockdown did not further increase fat-7 expression (Extended Data Fig. 3k). Deficiency in RSKS-1 or its upstream regulators (let-363/mTOR or daf-15/Raptor) all resulted in lifespan extension, and the ability of ash-2 or set-2 deficiency to extend lifespan was diminished in the context of rsks-1 deficiency (Extended Data Fig. 3o, p). Collectively, these data suggest that RSKS-1/S6K is a germline target of the H3K4me3 methyltransferase complex that mediates at least part of the intestinal upregulation of delta-9 desaturases leading to MUFA accumulation.
MUFAs mediate longevity by H3K4me3 modifiers
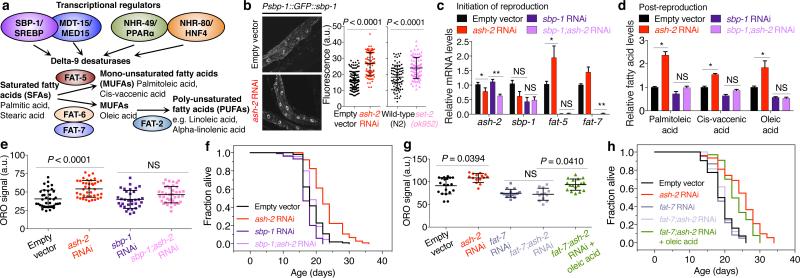
To examine the role of MUFAs in H3K4me3 modifiers-mediated longevity, we first further explored the molecular mechanisms underlying MUFA accumulation upon H3K4me3-methyltransferase deficiency. Delta-9 desaturases are known targets of a highly conserved intestinal transcriptional network, which involves the transcription factor SBP-1/SREBP19 and the Mediator complex subunit MDT-15/MED1520, as well as other transcription factors such as NHR-4921 and NHR-80/HNF422 (Fig. 4a). Knockdown of ash-2 and set-2, as well as set-2 mutation, increased nuclear accumulation of SBP-1::GFP in intestinal cells (Fig. 4b, Extended Data Fig. 4a), and this accumulation was reduced in rsks-1 deficient animals (Extended Data Fig. 4b-d). We next tested if SBP-1 (and MDT-15) is necessary for delta-9 desaturase upregulation and MUFA accumulation in H3K4me3-methyltransferase deficient worms. Knockdown of sbp-1 or mdt-15 decreased the basal levels of fat-5 and fat-7 mRNA and blunted their upregulation by ash-2 knockdown (Fig. 4c, Extended Data Fig. 4e). Knockdown of sbp-1 or mdt-15 also blocked the elevation of MUFAs (palmitoleic, cis-vaccenic, and oleic acid) in ash-2 deficient worms post-reproduction (Fig. 4d, Extended Data Fig. 4f). Thus, SBP-1/SREBP and MDT-15/MED15 are critical for delta-9 desaturase upregulation and MUFA accumulation in H3K4me3-methyltransferase deficient worms.
Figure 4. MUFA accumulation is critical for the longevity of H3K4me3-methyltransferase deficient worms.
a, Conserved transcriptional network regulating delta-9 desaturase genes. b, Images and quantification of SBP-1 nuclear accumulation. Mean ± s.d. of 2 independent experiments, each with n = 4-6 nuclei per worm of ≥ 8 worms per condition. c, RT-qPCR. Mean ± s.e.m. of 3 independent experiments, each with 3 biological replicates. d, GC-MS. Mean ± s.e.m. of 2 independent experiments, each with 2-3 biological replicates. e, ORO quantification. Mean ± s.d., n ≥ 32 worms per condition. f, Lifespan extension by ash-2 RNAi is reduced in sbp-1 RNAi (9.78%) compared to control worms (21.90%) (P = 0.0003, two-way ANOVA). g, As in e, n ≥ 13 worms per condition. h, Dietary oleic acid extends lifespan in the context of fat-7;ash-2 double RNAi (P < 0.0001, log-rank). g, h, Representative of 2 experiments. P values: b, Two-tailed Mann-Whitney; e, g, Kruskal-Wallis with Dunn's correction; c, d, Two-tailed Mann-Whitney with Benjamini-Hochberg correction. * P < 0.05, ** P < 0.01.
We next used this molecular knowledge to ask whether the MUFA metabolic switch is causative for the lifespan extension of H3K4me3-methyltransferase deficient worms. Interestingly, deficiency in sbp-1 or mdt-15 by knockdown or mutation abolished fat accumulation and lifespan extension in ash-2 knockdown worms (Fig. 4e, f, Extended Data Fig. 4g, h, Supplementary Table 1). Likewise, sbp-1 knockdown abrogated the longevity of set-2 mutant worms (Supplementary Table 1). In contrast, nhr-49 mutation only partially diminished the ability of ash-2 deficiency to promote fat accumulation and lifespan extension (Extended Data Fig. 4i, j) and nhr-80 mutation had no effect (Extended Data Fig. 4k, l). We also tested if the delta-9 desaturases themselves are required for the longevity of H3K4me3-methyltransferase deficient worms. Interestingly, fat-7 knockdown and fat-6;fat-7 double mutants, which are strongly deficient for oleic acid synthesis23, blocked the fat accumulation and longevity of ash-2 deficient worms (Fig. 4g, h, Extended Data Fig. 5a, b). In contrast, fat-7 and fat-6 hypomorphic single mutants (or fat-5;fat-7 and fat-5;fat-6 double mutants), which are only mildly deficient for MUFA synthesis22,23, did not abolish the longevity of ash-2 knockdown worms (Extended Data Fig. 5c-f). Thus, a conserved genetic pathway involving SBP-1/SREBP, MDT-15/MED15, and the downstream delta-9 desaturase FAT-7 is critical for the longevity of H3K4me3-methyltransferase deficient worms.
To more specifically assess the role of MUFAs in H3K4me3-methyltransferase deficient worms, we supplemented oleic acid in the diet (Extended Data Fig. 5g, h). Dietary oleic acid restored in large part the high fat phenotype and lifespan extension of ash-2;fat-7 double knockdown worms (Fig. 4g, h, Extended Data Fig. 5i, j). Similarly, dietary oleic acid restored high fat levels in ash-2;sbp-1 double knockdown worms (Extended Data Fig. 5k). Furthermore, knock-down of fat-2, which normally converts oleic acid into downstream PUFA13, increased fat accumulation and lifespan extension, and ash-2 deficiency did not further increase fat levels and extend lifespan in fat-2 knockdown worms (Extended Data Fig. 5l-o). Thus, the MUFA oleic acid plays a key role in the longevity of H3K4me3-methyltransferase deficient worms.
Dietary MUFAs extend lifespan
The role of oleic acid in lifespan regulation in the context of H3K4me3-methyltransferase deficiency led us to systematically examine the importance of MUFAs and their downstream PUFAs in lifespan regulation under physiological conditions (Fig. 5a). Dietary supplementation of individual MUFAs (oleic, palmitoleic, or cis-vaccenic acid) (Fig. 5b, d, Extended Data Fig. 6a) was sufficient to extend lifespan (Fig. 5c, e, f, Extended Data Fig. 6b). In contrast, dietary supplementation of the two PUFAs linoleic or alpha-linolenic acid (Extended Data Fig. 6c, e) did not significantly affect lifespan (Extended Data Fig. 6d, f, Fig. 5f). All MUFAs and PUFAs tested also increased total fat accumulation, except for the PUFA alpha-linolenic acid (Fig. 5c, e, Extended Data Fig. 6b, d, f). Furthermore, overexpressing the oleic acid-synthesizing enzyme FAT-7 in the intestine increased fat accumulation and extended lifespan (Fig. 5g, Extended Data Fig. 6g-i). Oleic acid supplementation did not further extend the long lifespan of FAT-7 overexpressing worms (Fig. 5g, Extended Data Fig. 6i). Collectively, these results suggest that dietary supplementation and endogenous accumulation of MUFAs, but not downstream PUFAs, are beneficial for lifespan.
Figure 5. Dietary supplementation of MUFAs is sufficient to extend lifespan.
a, Chemical structure of fatty acids used in supplementation experiments. b, GC-MS. Mean ± s.e.m. of 3 independent experiments, each with 2-3 biological replicates. c, Oleic acid supplementation extends lifespan in wild-type worms (P < 0.0001, log-rank). Inset: ORO quantification. Mean ± s.d., n ≥ 27 worms per condition. d, GC-MS. Mean ± s.e.m. of 2 independent experiments, each with 2-3 biological replicates. e, Palmitoleic acid supplementation extends lifespan in wild-type worms (P < 0.0001, log-rank). Inset: as in c, n ≥ 45 worms per condition. f, Mean lifespan changes in independent supplementation experiments (P values: log-rank). g, FAT-7 overexpression extends lifespan (P < 0.0001, log-rank), which is not further extended by dietary oleic acid. P values: Two-tailed Mann-Whitney (Benjamini-Hochberg correction for > 1 comparison). * P < 0.05, ** P < 0.01, *** P < 0.001.
Discussion
Our study shows that histone methylation modifiers influence MUFA metabolism. We propose a model whereby H3K4me3-methyltransferase deficiency causes the downregulation of specific targets (e.g. RSKS-1/S6K) in the germline, which in turn triggers a germline-to-intestine signal leading to SBP-1/SREBP activation, delta-9 desaturase expression, and MUFA accumulation in the intestine (Extended Data Fig. 6j). The nature of this germline-to-intestine signal is unknown, but it could be a diffusible lipid signal (‘lipokine’), a signal from the maturing eggs, or altered levels of lipid-binding proteins. Deficiency in the H3K4me3 methyltransferase complex might also affect fat metabolism by impacting the methyl pool available for other enzymatic reactions or by methylating yet-to-be-identified non-histone substrates.
Our work also shows that dietary MUFAs, but not their downstream PUFAs, positively impact lifespan. Previous studies have tested supplementation of individual fatty acids on lifespan24-28 and identified other PUFAs implicated in lifespan regulation24,26. Dietary oleic acid did not extend wild-type lifespan in previous studies25,27,28, although they used different oleic acid supplementation protocols. MUFAs could contribute to longevity by promoting membrane fluidity, minimizing oxidative stress, enhancing energy storage, or activating signaling pathways. The beneficial effects of MUFAs are exciting because of their dietary availability (e.g. oleic acid in olives) and microbiotic sources (e.g. palmitoleic acid in E. coli)29. In humans, diets rich in MUFAs have been associated with decreased risk of cardiovascular disease and diabetes30. The general conservation of lipid metabolism between worms and mammals raises the interesting possibility that the benefits of MUFAs on healthspan and lifespan could be conserved in other species.
Materials and Methods
Worm strains
All strains were maintained on standard nematode growth medium (NGM) plates with E. coli (OP50-1, streptomycin-resistant). Unless otherwise noted, worms were grown on the RNAi strain HT115 (DE3) for all experiments. The wild-type N2 strain (ABR) was provided by Dr. Man-Wah Tan. The NL2098 rrf-1(pk1417) strain, which restricts RNAi to the germline, intestine, and partially the hypodermal seam cells11,31, was provided by Andrew Fire. The germline and intestine RNAi strain AMJ345 rde-1(ne219); jamSi2[Pmex-5::rde-1(+)] and corresponding wild-type N2 (AMJ) were provided by Dr. Antony Jose10. The intestine specific RNAi strain VP303 rde-1(ne219); kbIs7[nhx-2p::rde-1 + rol-6(su1006)] was obtained from Caenorhabditis Genetics Center (CGC). Other strains obtained from CGC were as listed: RB1025 set-2(ok952), ZR1 rbr-2(tm1231), DA1116 eat-2(ad1116), XA7702 mdt-15(tm2182), STE68 nhr-49(nr2041), BX165 nhr-80(tm1011), BX153 fat-7(wa36), BX106 fat-6(tm331), BX160 fat-7(wa36);fat-5(tm420), BX110 fat-6(tm331);fat-5(tm420), BX156 fat-6(tm331);fat-7(wa36), RB1206 rsks-1(ok1255), and fluorescence translational fusion reporters BX150 lin-15B&lin-15A(n765); waEx18[fat-5::GFP + lin15(+)], BX113 lin-15B&lin-15A(n765); waEx15[fat-7::GFP + lin-15(+)], and CE548 sbp-1(ep79); epEx141[sbp-1::GFP::SBP-1 + rol-6(su1006)]. Mutants set-2(ok952), rbr-2(tm1231), eat-2(ad1116), and transgenic strain CE548 were outcrossed to our laboratory N2 (ABR) at least 6 times. During backcrossing, the sbp-1(ep79) mutation in CE548 was removed to generate a SBP-1::GFP translational fusion reporter in a wild-type background. See Supplementary Table 6 for a list of strains. All experiments were performed using hermaphrodites.
RNA interference
RNAi interference was performed as described6. HT115 (DE3) bacteria were transformed with vectors expressing dsRNA targeting the following genes of interest: ash-2, set-2, wdr-5.1, set-16, utx-1, jmjd-3.1, fat-2, fat-5, fat-7, mdt-15, sbp-1, tbh-1, nhr-10, F21A3.11, dod-23 (Ahringer Library, Dr. Andrew Fire), let-363/mTOR (Dr. Xiaomeng Long), daf-15/Raptor (Dr. Allen Hsu), rsks-1, asm-2 and unc-132 (Vidal Library, Dharmacon). All vectors were confirmed by sequencing. Empty vector bacteria, containing the RNAi plasmid without an RNAi insert, served as a control for all RNAi experiments. RNAi culture was concentrated 30-fold, and stored at 4°C for no more than 2 weeks. For double RNAi treatments, two cultures were mixed in a 1:1 ratio by volume. To obtain a synchronized worm population, egglay was performed on appropriate RNAi plates for 1-4 hours. All RNAi treatments began at hatching, except for ORO and lifespan experiments using sbp-1, mdt-15, fat-2, let-363/mTOR, or daf-15/Raptor RNAi, which began at initiation of reproduction (day 2.5 of life) to minimize effects on development.
Lifespan assays
Worm lifespan assays were performed at 20°C as described32. Kaplan-Meir lifespan curves were generated using GraphPad Prism, and statistical results were obtained by log-rank analysis using the JMP software. Two-way ANOVA was used to assess lifespan interactions between conditions. Lifespan assays that were blinded or repeated by an independent investigator are indicated in Supplementary Table 1. Representative Kaplan-Meir lifespan curves are shown in the figures and all statistics are included in Supplementary Table 1.
Oil Red O and Nile Red staining and quantification
Oil Red O (ORO) staining of fixed worms was conducted as described7, except that 2% Paraformaldehyde in PBS was used to fix worm tissues. ORO staining of dissected intestines was performed for one hour. Nile Red staining of fixed worms was performed as described33, except that staining was conducted in 1.5 mL Eppendorf tubes. ORO- or Nile Red-stained worms were mounted onto 2% agar pads, and imaged at 10x magnification using a Zeiss AxioSkop 2 Plus. Images were acquired using an AxioCam MRc camera with AxioVision 4.7 software and saved as TIF files. The same exposure settings were used across all conditions within each experiment. Using FIJI image processing software, raw images were background-subtracted, grayscale-converted, inverted, and thresholded to outline the worm body. The same threshold values were used across all conditions within each experiment. “Analyze Particles” was used to measure mean ORO intensity per worm. To quantify anterior intestinal cells of whole worms or extruded intestines of dissected worms, specific regions in the raw images were selected using the oval brush tools (intestine cells) or the segmented line tool (extruded intestines), saved to the Region of Interest (ROI) manager, processed as described above, and quantified for mean ORO intensity per selected region. Mean intensity values in arbitrary units (a.u.) were graphed using Graphpad Prism, and statistical significance was determined using a two-tailed Mann-Whitney test (2 conditions) or the Kruskal-Wallis test with Dunn's correction for multiple comparisons (more than 2 conditions). For figures containing straightened whole worms or intestines, unmodified images were selected and straightened, and representative worms or intestines from each condition were concatenated into montages.
GFP quantification
For FAT-5::GFP and FAT-7::GFP fluorescence reporters, worms at active reproduction (day 3 of life) were anesthetized in M9 (22 mM KH2PO4, 42 mM Na2HPO4, 86 mM NaCl, 1mM MgSO4, dissolved in water) containing100 μM levamisole and mounted on 2% agar pads. All images were acquired on Zeiss AxioSkop 2 Plus using an AxioCam MRc camera with AxioVision 4.7 software with identical exposure settings for the same experiment. Mean fluorescence intensity in arbitrary units (a.u.) of the whole worm was quantified in FIJI as described above. These experiments were conducted in a blinded manner. For visualization of intestinal nuclear SBP-1::GFP expression, worms at active reproduction (day 3 of life) were mounted on 2% agarose pads and anesthetized with 20 mM tetramisole in M9. Images were acquired using Zeiss Axio Imager.M2 microscope equipped with an ApoTome.2 system and AxioCam MRc camera using identical exposure settings for the same experiment. Images showing the maximum number of defined intestine nuclei were selected in each animal and saved as TIF files. Mean fluorescence intensity in arbitrary units (a.u.) of each nucleus was quantified using the oval brush tool in FIJI as described above.
Worm tissue micro-dissection
Synchronized worms grown on appropriate RNAi bacteria were collected at specified stages of life. At least 20 worms from each condition were washed 3 times with M9 buffer and deposited onto a glass slide in drops of M9 (one worm per drop) with a glass Pasteur pipette. Under the dissection light microscope, individual worms were decapitated using 22.5° knives (Surgistar) to extrude their anterior intestines and full germlines (including developing oocytes). For ORO or Nile Red staining, reproductive worms (day 5 of life) with extruded intestines were used for staining. For RT-qPCR or RNA-seq, fully detached intestines and germlines from worms during active reproduction (day 3 of life) or at initiation of reproduction (day 2.5 of life) were harvested. To isolate the full germline containing maturing oocytes, an incision was made before the spermatheca to minimize sperm contamination. Dissected germlines were immediately collected in 500 μL of Trizol LS reagent (Invitrogen) using a glass Pasteur pipet whose tip was pulled under the flame to thin out the opening. To isolate the anterior intestine from the same worms, an incision was made near the anterior 10 pairs of intestinal cells. The anterior intestine samples were collected in Trizol in separate tubes. All samples were stored at −80°C until RNA extraction.
Quantitative RT-PCR
For experiments using whole worms, at least 100 aged-synchronized worms in biological triplicate for each condition were harvested, washed 3 times with M9, and resuspended in 500 μL of Trizol. For experiments using dissected tissues, 20-40 germlines or intestines in biological triplicate for each condition were harvested. To extract total RNA, worm or tissue pellets in Trizol underwent six freeze-thaw cycles in a dry ice-ethanol bath. RNA was extracted according to the Trizol procedure, and resuspended in RNase- and DNase-free water. RNA from whole worms was quantified using the Qubit 2.0 fluorometer (Invitrogen), and at least 500 ng of total RNA per condition was used for cDNA synthesis. RNA from dissected tissues was quality-assessed and quantified using the Agilent Bioanalyzer (RNA 6000 Pico), and at least 2 ng of total RNA per condition was used for cDNA synthesis.
Total RNA was treated with DNAse I Amp Grade (Invitrogen), then reverse-transcribed using Oligo-dT primers (Invitrogen) and Superscript III reverse transcriptase (Invitrogen), based on the manufacturer's instructions. RT-qPCR was performed using diluted cDNA on an ABI7900HT or a Bio-Rad C1000 thermal cycler using iTaq master mix (Bio-Rad) containing a passive reference dye (ROX™). Primers used in RT-qPCR were designed to span exon-exon junctions towards the 3’ end of the gene and used at a final concentration of 250 nM each. Primers are listed in Supplementary Table 7. Melt curves were examined to ensure primer specificity. Results were analyzed using the standard ΔΔCT method. For each biological replicate, the mean CT value of 3-4 technical replicates was analyzed. act-1 served as the reference gene in all analyses, and changes in mRNA levels relative to act-1 were confirmed using either cdc-42, ama-1, or pmp-3 as alternate reference genes in independent experiments (data not shown).
RNA-seq on micro-dissected tissues
For each of the five replicates, full germlines and anterior intestines were collected from 20 worms treated with ash-2 RNAi or empty vector control. At the initiation of reproduction (day 2.5 of life), worms from each pair of ash-2 RNAi and empty vector plates were dissected within the same hour. Tissue dissection and RNA extraction procedures were the same as described above and 20 full germlines or ~200 intestinal cells were used as initial input for individual RNA-seq libraries. For cDNA synthesis, 150 pg (intestine) or 750 pg (germline) of extracted RNA was treated with RQ1 RNase-free DNase (Promega), which was later removed, and reverse transcribed using oligo(dT)-primed, Clontech SMART-Seq v4 Ultra Low Input RNA Kit (634889) according to manufacturer's instructions. Poly-A selected RNA-seq libraries were generated using Illumina Nextera XT DNA Library Prep Kit (FC-131-1024). All 20 libraries were processed together to minimize batch effects. The libraries were uniquely barcoded and multiplexed onto one lane, and 150 base pair paired-end reads were sequenced on NextSeq 500 (Illumina).
RNA-seq analysis
RNA-seq reads were aligned to the WBcel235 genome and gene read counts were calculated using STAR v2.5.1b34. Low-coverage genes were filtered out: only genes with at least 1 read count per million mapped reads in at least 5 samples were included in the analysis, resulting in a dataset of 10,442 genes and 20 samples. More genes were detected in the germline samples (185 genes with a median count number of 0) than in the intestine samples (682 genes with a median count number of 0), with 76% of the genes having a higher median read count in germline samples than in intestine samples. Because of these tissue-specific differences, we did not perform an across-sample normalization (which assumes that most genes are not differentially expressed) and instead used FPKM values to compare germline and intestine samples. The read counts were transformed to log2(FPKM+1) values, which were used for the principal component analysis (PCA) on the samples from both tissues together. A gene was determined to be germline-enriched if the lowest expression value (log2(FPKM+1)) in the germline empty-vector samples was at least 2-fold higher than the highest expression value in the intestine empty-vector samples (5,494 germline-enriched genes). A gene was determined to be intestine-enriched if the lowest expression value (log2(FPKM+1)) in the intestine empty-vector samples was at least 2-fold higher than the highest expression value in the germline empty-vector samples (1,418 intestine-enriched genes).
We then compared empty-vector and ash-2 RNAi samples for each tissue separately. Low-coverage genes were filtered out: only genes with at least 1 read count per million mapped reads in at least 5 samples were included in the analysis. This resulted in a germline dataset of 9,073 genes and 10 samples, and an intestine dataset of 7,288 genes and 10 samples. Using DESeq2 v1.10.135, the read counts were normalized using the variance stabilizing transformation (VST) within each tissue and PCA was performed on the germline and intestine dataset. For each tissue, differentially expressed genes between empty-vector and ash-2 RNAi were examined, accounting for a plate effect (GE ~ condition + plate). Gene-wise P values were corrected for multiple hypothesis testing using Benjamini-Hochberg and FDR = 0.05 was used as a significance threshold.
Selection of ASH-2 candidate targets
ASH-2 candidate targets were selected based on the following datasets: 1) our RNA-seq dataset, 2) microarrays on ash-2 knockdown larval worms6 and ash-2 mutant adult germlines14, and 3) ASH-2 ChIP-chip on worm embryos15. Predicted candidate expression in germline/intestine for datasets 2) and 3) was based on published gene expression datasets36-38 (Andreas Rechtsteiner and Susan Strome, personal communication) and pre-publication RNA-seq datasets (Robert Waterston, personal communication). The following criteria were used to select potential targets: i) downregulation upon ash-2 deficiency, as the H3K4me3 methyltransferase complex is generally associated with gene activation; ii) expression in the germline or intestine, as these are the two key tissues involved in this fat metabolic switch; and/or iii) previously identified role in lifespan or fat metabolism.
Generation of transgenic plasmids
The Pges-1::fat-7 plasmid (pSH1) contains 2kb of ges-1 promoter region (Pges-1) upstream of fat-7 full genomic DNA sequence followed by fat-7 3’ UTR in the P4-P1R vector. The plasmid containing Pges-1 in the P4-P1R vector was a gift from Dr. Frederick Mann. The fragment containing fat-7 full genomic DNA (containing the coding region and 3’UTR) was generated by PCR amplification from wild-type (N2) genomic DNA using the following primers (pSH1_fat-7_F: 5’-CTATTACATATCTTATCTTTGAATTCAGATGACGGTAAAAACTCGCGCGAGCATTGCCAA-3’ and pSH1_fat-7_R: 5’-GCCGCCCTGCAGCTCTAGAGCTCGAATTCTTAGAATTAACCAAATTTATTCAGGAATAA-3’). The fragment containing Pges-1 and the P4-P1R vector was PCR-amplified using the following primers (pSH1_Pges-1 _F: 5’-TTGGCAATGCTCGCGCGAGTTTTTACCGTCATCTGAATTCAAAGATAAGATATGTAATAG-3’and pSH1_Pges-1_R: 5’-TTTATTCCTGAATAAATTTGGTTAATTCTAAGAATTCGAGCTCTAGAGCTGCAGGGCGGC-3’). The fragments were assembled via Gibson assembly39.
The Pges-1::set-2a cDNA::SL2::GFP plasmid (pSH9) contains 2kb of ges-1 promoter region (Pges-1) upstream of the full-length cDNA of set-2.a (the long isoform of set-2) followed by the SL2 trans-splicing sequence, GFP cDNA, and unc-54 3’UTR in the pIM26 vector. pIM26 containing SL2 upstream of GFP and unc-54 3’UTR was a gift from Yue Zhang. The 2kb Pges-1 fragment upstream of ges-1 ATG was generated by PCR amplification from wild-type (N2) genomic DNA using the following primers (pSH9_ Pges-1_F: 5’-ATGCCTGCAGGTCGACTCTAGAGGATCCCCAAACTCCGAACTATGATGACGAAAAAATGT-3’ and pSH9_Pges-1_R: 5’-AGGATGATGGTTCATATCATGTGTGGACATCTGAATTCAAAGATAAGATATGTAATAGAT-3’). The set-2.a cDNA fragment was generated by PCR amplification from pAS2.1 (a gift from Dr. Francesca Palladino) using the following primers (pSH9_set-2.a_F: 5’-ATCTATTACATATCTTATCTTTGAATTCAGATGTCCACACATGATATGAACCATCATCCT- 3’ and pSH9_set-2.a_R: 5’-AACTAGGTGAAAGTAGGATGAGACAGCCCCTCAATTAAGATATCCACGACACGTCTTCGC- 3’), and was corrected for a short missing sequence. The pIM26 fragment was amplified by PCR using the following primers (pSH9_pIM26_F: 5’-GCGAAGACGTGTCGTGGATATCTTAATTGAGGGGCTGTCTCATCCTACTTTCACCTAGTT -3’and pSH9_pIM26_R: 5’-ACATTTTTTCGTCATCATAGTTCGGAGTTTGGGGATCCTCTAGAGTCGACCTGCAGGCAT-3’). Three fragments were assembled via Gibson assembly39.
All fragments generated via PCR amplification were sequence-verified in the final plasmid to ensure no mutations were introduced.
Generation of transgenic worm strains
The strain carrying extra-chromosomal arrays of [Pges-1::fat-7; myo-3::mCherry] was generated by injecting wild-type N2 (ABR) worms with pSH1 and a co-injection plasmid pCFJ104, with a muscle specific myo-3 promoter driving the mCherry gene (Pmyo-3::mCherry, made by Dr. Erik Jorgensen, Addgene plasmid #19328). The strain carrying extra-chromosomal arrays of [Pges-1::set-2a cDNA; myo-3::mCherry] was generated by injecting set-2(ok952) (ABR10) worms with pSH9 and Pmyo-3::mCherry. The injection mixture of both plasmids was at a final concentration of 50 ng/μL. Control strains were generated by injecting wild-type N2 (ABR) worms with the co-injection marker (Pmyo-3::mCherry) at final a concentration of 50 ng/μL.
Worms containing extra-chromosomal arrays were selected based on mCherry fluorescence in the body wall muscles. In transgenic worms also carrying a GFP reporter, GFP fluorescence was used as a primary marker for strain maintenance. Overexpression of fat-7 in wild-type transgenic worms was confirmed by RT-qPCR with primers designed to detect fat-7 mRNA. Overexpression of set-2.a in set-2(ok952) mutant transgenic worms was confirmed by RT-qPCR with primers designed to detect transgenic (wild-type) but not mutant set-2.a mRNA. Functional validation of set-2 intestine rescue transgenic lines was conducted via RT-qPCR analysis of intestinal targets of COMPASS H3K4me3 complex, dod-23 and asm-2, which are normally downregulated in set-2(ok952) mutants. Primer sequences are listed in Supplementary Table 7.
GC-MS quantification of fatty acids
Quantification of total triglycerides and total phospholipids levels and profiling of long-chain fatty acids were performed as described33 with the following modifications. For each condition, 500-1000 age-synchronized animals were used. For GC-MS, an Agilent 7890A gas chromatograph equipped with an HP-5MS column (30 m × 0.25 μm I.D. × 0.25 μm, Agilent) using helium as a carrier gas was used, and 1 μm of each sample was injected at a 10:1 split ratio. The initial oven temperature was set to 150°C, increased to 220°C at 20°C/min, increased to 230°C at 2°C/min, increased to 320°C at 50°C/min, and held for 2 minutes at this temperature. Fatty acid abundance was expressed as a ratio (μg/mg) of fatty acid concentration (μg/mL) to protein concentration (mg/mL) for each sample. In all GC-MS figure panels involving comparisons to the empty vector control (unless otherwise mentioned), raw fatty acid concentration (μg/mg) of each condition was normalized to raw fatty acid concentration (μg/mg) of the empty vector control. The final ratio is expressed as relative fatty acid levels. Raw GC-MS data for all figure panels are provided in Supplementary Table 8.
Fertility assessment
Fertility of N2 worms treated with ash-2 RNAi or empty vector control was assayed at 20°C. After a one-hour egglay, and eggs were maintained at 20°C until worms reached initiation of reproduction (2.5 day of life). Each of 30 reproductive worms from each condition was transferred onto individual 3.5 cm plate seeded with corresponding bacteria. Individual worms were transferred onto a new plate every day, and the number of fertilized eggs (eggs with visible shell) and unfertilized oocytes (eggs without visible shell) were assessed on the old plate. The old plate with eggs was incubated at 20°C for an additional day, and the number of hatched progeny (live brood size) was counted. Fertility was assessed daily until no more hatched worms, fertilized eggs, or unfertilized oocytes were seen on each plate. This experiment was conducted in a blinded manner.
Fatty acid supplementation
The fatty acid supplementation protocol was adapted from40 with the following modifications. To facilitate fatty acid dissolution, the detergent Tergitol (type NP-40, Sigma-Aldrich) was added to a final concentration of 0.001% in liquid NGM agar media for both non-supplemented and supplemented plates prior to autoclaving. NGM plates for RNAi experiments were prepared as described6, and HT115 bacteria were used for all supplementation experiments. A final concentration ranging from 0.1 mM to 4 mM of oleic acid was used during the initial experimental optimization, and a final concentration of 0.8 mM was used for all fatty acids in subsequent supplementation experiments. HT115 bacteria expressing empty vector or the appropriate RNAi were seeded onto plates containing the respective fatty acids at room temperature for 24-48 hours before the addition of worms to ensure incorporation of fatty acids into feeding bacteria. Successful incorporation of supplemented fatty acids into the HT115 bacteria and into the worms was confirmed by GC-MS.
Data availability statement
RNA-seq data are available at Sequence Read Archive (SRA) under accession number PRJNA343151. The code for RNA-seq analysis is available on the Brunet Lab Github: https://github.com/brunetlab.
Extended Data
Extended Data Figure 1. Deficiency in H3K4me3 modifiers leads to fat accumulation in the intestine without impacting fertility.
a, ORO quantification. Mean ± s.d., n ≥ 17 worms per condition. b, ORO images. Scale bar: 100 μm. c, ORO quantification. Mean ± s.d., n ≥ 9 dissected intestines per condition. d-e, Nile Red staining and quantification. Mean ± s.d., n ≥ 7 dissected intestines (d) or n ≥ 11 worms (e) per condition. Scale bar: 100 μm. f, Autofluorescence and quantification. Mean ± s.d., n ≥ 17 worms per condition. Scale bar: 100 μm. g, Fertility quantification of live brood size (i), fertilized eggs (ii), and unfertilized oocytes (iii) laid per worm. Mean ± s.d., n ≥ 25 worms per condition. h, RT-qPCR. Mean ± s.e.m. of 2 independent experiments, each with 2-3 biological replicates. i, RT-qPCR. Mean ± s.e.m. from 2 independent experiments, each with 2-3 biological replicates. j, ORO quantification. Mean ± s.d., n ≥ 26 worms per condition. k, ORO quantification. Mean ± s.d., n ≥ 29 worms per condition. l, RT-qPCR. Mean ± s.e.m. of 3 biological replicates. m, D.I.C. (Nomarski) and GFP fluorescence images. n, ORO quantification. Mean ± s.d., n ≥ 26 worms per condition. a-f, k (rescue line #1), n, Representative of 2 experiments. P values: a, c-g, Two-tailed Mann-Whitney; h-i, Two-tailed Mann-Whitney with Benjamini-Hochberg correction; j-k, n, Kruskal-Wallis with Dunn's correction; l, Two-tailed t-test with Benjamini-Hochberg correction. * P < 0.05, ** P < 0.01.
Extended Data Figure 2. The delta-9 desaturases FAT-5 and FAT-7 support MUFA accumulation in H3K4me3-methyltransferase deficient worms.
a, GC-MS quantification of MUFAs. Mean ± s.e.m. of 2 independent experiments, each with 3 biological replicates. b-c, RT-qPCR. Mean ± s.e.m. of 2 independent experiments, each with 3 biological replicates. d, GC-MS quantification of MUFAs. Mean ± s.e.m. of 2 independent experiments, each with 2-3 biological replicates. e, RT-qPCR. Mean ± s.e.m. of 2 independent experiments, each with 3 biological replicates. P values: a-e, Two-tailed Mann-Whitney with Benjamini-Hochberg correction. * P < 0.05, ** P < 0.01.
Extended Data Figure 3. RNA-seq on micro-dissected germlines and intestines and functional validation of ASH-2 targets.
a, RNA-seq tissue sample collection pipeline. b, Principal component analysis (PCA) with both intestine and germline samples (left), only intestine samples (middle), or only germline samples (right). c, RT-qPCR. Mean ± s.e.m. of 2 independent experiments, each with 2-3 biological replicates. d-g, ORO quantification. Mean ± s.d., n ≥ 15 worms per condition. h, RT-qPCR. Mean ± s.e.m. of 2 independent experiments, each with 3 biological replicates. i-j, ORO quantification. Mean ± s.d., n ≥ 21 (i) and n ≥ 27 (j) worms per condition. k, RT-qPCR. Mean ± s.e.m. of 2 independent experiments, each with 3 biological replicates. l, GC-MS. Mean ± s.e.m. of 2 independent experiments, each with 3 biological replicates. m, ORO quantification. Mean ± s.d., n ≥ 19 worms per condition. n, RT-qPCR. Mean ± s.e.m. of 2 independent experiments, each with 3 biological replicates. o, Lifespan extension by ash-2 RNAi is reduced in rsks-1(ok1255) mutants (13.12%) compared to wild-type worms (29.20%) (P < 0.0001, two-way ANOVA). p, let-363/mTOR RNAi and daf-15/Raptor RNAi extend lifespan in wild-type worms (P < 0.0001, log-rank), but not in set-2(ok952) mutants. d (except rrf-1 data), i-j, Representative of 2 experiments. P values: c, k, l, n, Two-tailed Mann-Whitney with Benjamini-Hochberg correction; h, Two-tailed Mann-Whitney; d-g, i-j, m, Kruskal-Wallis with Dunn's correction. * P < 0.05, ** P < 0.01.
Extended Data Figure 4. Role of SBP-1/SREBP, MDT-15/MED15, NHR-49, and NHR-80 in the fat accumulation and longevity of H3K4me3-methyltransferase deficient worms.
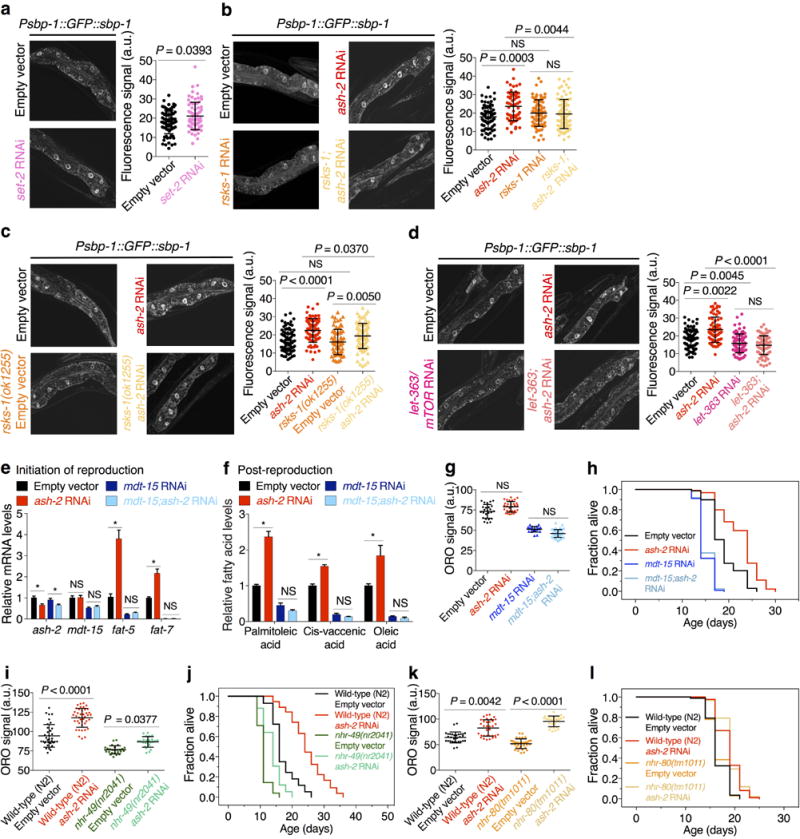
a-d, Images and quantification of SBP-1 nuclear accumulation. Mean ± s.d. of 2 independent experiments, each with 4-6 nuclei per worm of ≥ 8 worms per condition. e, RT-qPCR. Mean ± s.e.m. of 2 independent experiments, each with 3 biological replicates. f, GC-MS quantification of MUFAs. Mean ± s.e.m. of 2 independent experiments, each with 2-3 biological replicates. g, ORO quantification. Mean ± s.d., n ≥ 21 worms per condition. h, ash-2 RNAi extends lifespan in wild-type worms (P < 0.0001, log-rank), but not in mdt-15 RNAi worms. i, ORO quantification, Mean ± s.d., n ≥ 24 worms per condition. j, ash-2 RNAi extends lifespan in both wild-type worms and nhr-49(nr2041) mutants (P < 0.0001, log-rank). k, ORO quantification. Mean ± s.d., n ≥ 27 worms per condition. l, ash-2 RNAi extends lifespan in both wild-type worms and nhr-80(tm1011) mutants (P < 0.0001, log-rank). i-k, Representative of 2 experiments. P values: a, Two-tailed Mann-Whitney; b-d, g, i, k, Kruskal-Wallis with Dunn's correction; e-f, Two-tailed Mann-Whitney with Benjamini-Hochberg correction. * P < 0.05.
Extended Data Figure 5. Delta-9 desaturases FAT-6 and FAT-7 and MUFA oleic acid mediate the longevity of H3K4me3-methyltransferase deficient worms.
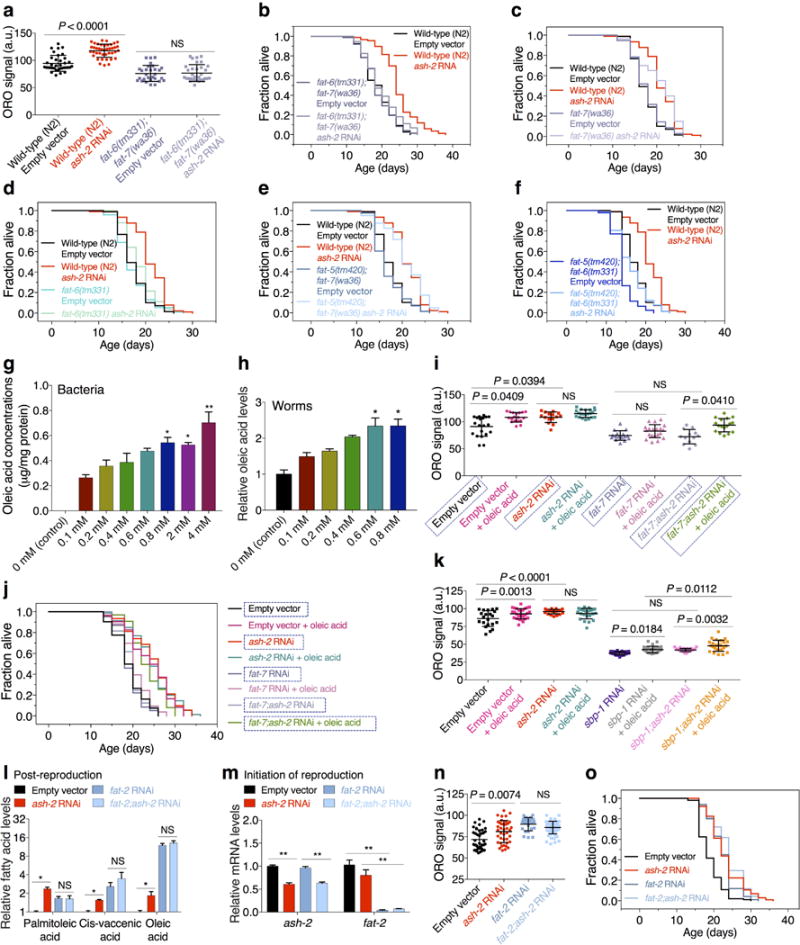
a, ORO quantification. Mean ± s.d., n ≥ 29 worms per condition. b, ash-2 RNAi leads to lifespan extension in control (P < 0.0001, log-rank), but not in fat-6(tm331);fat-7(wa36) mutants. c, ash-2 RNAi extends lifespan in both wild-type worms and fat-7(wa36) single mutants (P < 0.0001, log-rank). d, ash-2 RNAi extends lifespan in fat-6(tm331) single mutants (P = 0.0162, log-rank), but lifespan extension by ash-2 RNAi is reduced in fat-6(tm331) mutants (9.26%) compared to wild-type worms (20.46%) (P = 0.0072, two-way ANOVA). e, ash-2 RNAi extends lifespan in fat-5(tm420);fat-7(wa36) double mutants (P < 0.0001, log-rank). f, ash-2 RNAi extends lifespan in fat-5(tm420);fat-6(tm331) worms (P = 0.0002, log-rank), but lifespan extension by ash-2 RNAi is reduced in fat-5(tm420);fat-6(tm331) mutants (14.03%) compared to wild-type worms (20.46%) (P = 0.0358, two-way ANOVA). g-h, GC-MS quantification of oleic acid. Mean ± s.e.m. of 3 biological replicates. i, ORO quantification. Mean ± s.d., n ≥ 13 worms per condition. Boxed conditions are identical to Fig. 4g. j, Oleic acid supplementation extends lifespan (P < 0.0001, log-rank), which is not further extended by ash-2 RNAi. Oleic acid supplementation extends lifespan in fat-7;ash-2 RNAi worms (P < 0.0001, log-rank). Boxed conditions are identical to Fig. 4h. k, ORO quantification. Mean ± s.d., n ≥ 21 worms per condition. l, GC-MS quantification of MUFAs. Mean ± s.e.m. of 2 independent experiments, each with 2-3 biological replicates. m, RT-qPCR. Mean ± s.e.m. of 2 independent experiments, each with 3 biological replicates. n, ORO quantification. Mean ± s.d., n ≥ 25 worms per condition. o, ash-2 RNAi extends lifespan in control (P < 0.0001, log-rank) but not fat-2 RNAi worms. c, i-j, n-o, Representative of 2 experiments. P values: a, g-i, k, n, Kruskal-Wallis with Dunn's correction; l-m, Two-tailed Mann-Whitney with Benjamini-Hochberg correction. * P < 0.05, ** P < 0.01.
Extended Data Figure 6. Dietary supplementation of MUFAs, but not PUFAs, extends lifespan in wild-type worms.
a, GC-MS. Mean ± s.e.m. of 2 independent experiments, each with 2-3 biological replicates. b, Cis-vaccenic acid supplementation extends lifespan in wild-type worms (P < 0.0001, log-rank). Inset: ORO quantification. Mean ± s.d., n ≥ 20 worms per condition. c, GC-MS. Mean ± s.e.m. of 2 independent experiments, each with 3 biological replicates. d, Linoleic acid supplementation does not extend lifespan in wild-type worms. Inset: ORO quantification. Mean ± s.d., n ≥ 23 worms per condition. e, GC-MS. Mean ± s.e.m. of 2 independent experiments, each with 3 biological replicates. f, Alpha-linolenic acid supplementation does not increase the lifespan of wild-type worms. Inset: ORO quantification. Mean ± s.d., n ≥ 23 worms per condition. g, RT-qPCR. Mean ± s.e.m. of 3 biological replicates. h, ORO quantification. Mean ± s.d., n ≥ 54 worms per condition. i, Overexpression of FAT-7 extends lifespan (P < 0.0001, log-rank), which is not further extended by dietary oleic acid. Boxed regions are identical to Fig. 5g. j, Proposed model by which ash-2 deficiency in the germline could lead to the fat metabolic switch in the intestine. b, d, f, Representative of 2 experiments. P values: a, c, e, Two-tailed Mann-Whitney with Benjamini-Hochberg correction; b inset, d inset, f inset, Two-tailed Mann-Whitney; g, Kruskal-Wallis test with Dunn's correction (non-significant, likely due to small sample size). One-way ANOVA with Bonferroni's correction. h, Kruskal-Wallis with Dunn's correction; * P < 0.05, ** P < 0.01, *** P < 0.001.
Extended Data Table 1. List of ASH-2 candidate targets.
List of candidate targets of ASH-2 based on our gene expression dataset as well as published datasets.
| Candidates | Datasets | Functions | Expression |
|---|---|---|---|
| tbh-1 | RNA-seq of ash-2 knockdown worms (adult, micro-dissected tissues) | Tyramine beta-hydroxylase, Required for biosynthesis of the octopamine neurotransmitter, Longevity | Germline |
| nhr-10 | RNA-seq of ash-2 knockdown worms (adult, micro-dissected tissues) | Nuclear hormone receptor, Fat metabolism | Intestine, Germline |
| unc-132 | RNA-seq of ash-2 knockdown worms (adult, micro-dissected tissues) | Unknown roles in fat metabolism and longevity | Germline |
| F21A3.11 | RNA-seq of ash-2 knockdown worms (adult, micro-dissected tissues) | Predicted to have acid phosphatase activity and metal ion binding activity, Longevity | Intestine |
| asm-2 | RNA-seq of ash-2 knockdown worms (adult, micro-dissected tissues) Microarray of ash-2 knockdown worms (larval, whole worms) |
Acid sphingomyelinase | Intestine |
| dod-23 | RNA-seq of ash-2 knockdown worms (adult, micro-dissected tissues) Microarray of ash-2 knockdown worms (larval, whole worms) |
Determinant of adult lifespan | Intestine |
| acs-2 | Microarray of ash-2 knockdown worms (larval, whole worms) | Fatty acid beta-oxidation | Intestine (predicted) |
| acdh-2 | Microarray of ash-2 knockdown worms (larval, whole worms) | Fatty acid beta-oxidation | Intestine (predicted) |
| rsks-1 | ASH-2 ChIP-chip (whole embryos) | S6 kinase (S6K), Fat metabolism, Longevity | Germline (predicted) |
| aak-2 | ASH-2 ChIP-chip (whole embryos) | AMPK subunit, Nutrient sensing, Longevity | Intestine, Germline (predicted) |
Supplementary Material
Acknowledgements
We are grateful to A. Jose, A. Rechtsteiner, S. Strome, and R. Waterston, for sharing expression data and strains pre-publication. We thank A. Fire, M. Hansen, S. Kim, F. Palladino, D. Pattabiraman, Y. Zhang, and the Caenorhabditis Genetics Center for plasmids and strains. We thank M. Hansen, E. O'Rourke, L. Booth, C-K. Hu, D. Leeman, J. Lim, and S. Mahmoudi for critically reading the manuscript, and A. Fire, O. Gozani, S. Kim, and Brunet lab members for helpful discussions. We thank A. Chien for GC-MS consulting and J. Coller at the Stanford Functional Genomics Facility. Supported by NIH DP1AG044848 (A.B.), NIH R01AG054201 (A.B. and W.B.M.), NIH R01AG044346 (W.B.M.), a Stanford Mass Spectrometry grant (S.H. and A.B.), NSF Graduate Research Fellowship, Stanford Graduate Fellowship, and NIH T32AG047126 (S.H.), and NIH T32AG047126 and NIH F32AG051337 (E.A.S.).
Footnotes
Author Contributions
S.H. conceived the study under the guidance of A.B.. S.H. performed all the experiments except those specified below. E.A.S. planned and performed Nile Red and FAT-5/FAT-7 reporter imaging, time-course RT-qPCR, and one oleic acid supplementation lifespan experiment. C.G.S.G. planned and performed SBP-1 localization experiments and generated set-2 transgenic lines under the guidance of W.B.M.. K.H. analyzed the RNA-seq data. S.H. wrote the paper with the help of A.B. and E.A.S.. C.G.S.G., K.H., and W.B.M. provided feedback.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Supplementary Information
Supplementary information is available in the online version of this paper.
References
- 1.Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol. 2015;16:593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sen P, Shah PP, Nativio R, Berger SL. Epigenetic Mechanisms of Longevity and Aging. Cell. 2016;166:822–39. doi: 10.1016/j.cell.2016.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller M, et al. Triglycerides and cardiovascular disease: a scientific statement from the American Heart Association. Circulation. 2011;123:2292–333. doi: 10.1161/CIR.0b013e3182160726. [DOI] [PubMed] [Google Scholar]
- 4.Hansen M, Flatt T, Aguilaniu H. Reproduction, fat metabolism, and life span: what is the connection? Cell Metab. 2013;17:10–9. doi: 10.1016/j.cmet.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greer EL, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–7. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Rourke EJ, Soukas AA, Carr CE, Ruvkun GC. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–5. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks KK, Liang B, Watts JL. The influence of bacterial diet on fat storage in C. elegans. PLoS One. 2009;4:e7545. doi: 10.1371/journal.pone.0007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espelt MV, Estevez AY, Yin X, Strange K. Oscillatory Ca2+ signaling in the isolated Caenorhabditis elegans intestine: role of the inositol-1,4,5-trisphosphate receptor and phospholipases C beta and gamma. J Gen Physiol. 2005;126:379–92. doi: 10.1085/jgp.200509355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marré J, Traver EC, Jose AM. Extracellular RNA is transported from one generation to the next in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2016;113:12496–12501. doi: 10.1073/pnas.1608959113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumsta C, Hansen MC. elegans rrf-1 mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS One. 2012;7:e35428. doi: 10.1371/journal.pone.0035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watts JL, Browse J. A palmitoyl-CoA-specific delta9 fatty acid desaturase from Caenorhabditis elegans. Biochem Biophys Res Commun. 2000;272:263–9. doi: 10.1006/bbrc.2000.2772. [DOI] [PubMed] [Google Scholar]
- 13.Peyou-Ndi MM, Watts JL, Browse J. Identification and characterization of an animal delta(12) fatty acid desaturase gene by heterologous expression in Saccharomyces cerevisiae. Arch Biochem Biophys. 2000;376:399–408. doi: 10.1006/abbi.2000.1733. [DOI] [PubMed] [Google Scholar]
- 14.Robert VJ, et al. The SET-2/SET1 histone H3K4 methyltransferase maintains pluripotency in the Caenorhabditis elegans germline. Cell Rep. 2014;9:443–50. doi: 10.1016/j.celrep.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 15.Pferdehirt RR, Kruesi WS, Meyer BJ. An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes Dev. 2011;25:499–515. doi: 10.1101/gad.2016011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapahi P, et al. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–65. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi X, et al. Regulation of lipid droplet size and phospholipid composition by stearoyl-CoA desaturase. J Lipid Res. 2013;54:2504–14. doi: 10.1194/jlr.M039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang F, et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–4. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 20.Taubert S, Van Gilst MR, Hansen M, Yamamoto KR. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 2006;20:1137–49. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 2005;3:e53. doi: 10.1371/journal.pbio.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brock TJ, Browse J, Watts JL. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet. 2006;2:e108. doi: 10.1371/journal.pgen.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brock TJ, Browse J, Watts JL. Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans. Genetics. 2007;176:865–75. doi: 10.1534/genetics.107.071860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shmookler Reis RJ, et al. Modulation of lipid biosynthesis contributes to stress resistance and longevity of C. elegans mutants. Aging (Albany NY) 2011;3:125–47. doi: 10.18632/aging.100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goudeau J, et al. Fatty acid desaturation links germ cell loss to longevity through NHR-80/HNF4 in C. elegans. PLoS Biol. 2011;9:e1000599. doi: 10.1371/journal.pbio.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Rourke EJ, Kuballa P, Xavier R, Ruvkun G. omega-6 Polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev. 2013;27:429–40. doi: 10.1101/gad.205294.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratnappan R, et al. Germline signals deploy NHR-49 to modulate fatty-acid beta-oxidation and desaturation in somatic tissues of C. elegans. PLoS Genet. 2014;10:e1004829. doi: 10.1371/journal.pgen.1004829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee D, et al. SREBP and MDT-15 protect C. elegans from glucose-induced accelerated aging by preventing accumulation of saturated fat. Genes Dev. 2015;29:2490–503. doi: 10.1101/gad.266304.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magnuson K, Jackowski S, Rock CO, Cronan JE., Jr. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev. 1993;57:522–42. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gillingham LG, Harris-Janz S, Jones PJ. Dietary monounsaturated fatty acids are protective against metabolic syndrome and cardiovascular disease risk factors. Lipids. 2011;46:209–28. doi: 10.1007/s11745-010-3524-y. [DOI] [PubMed] [Google Scholar]
Materials and Methods References
- 31.Sijen T, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–76. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 32.Greer EL, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17:1646–56. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pino EC, Webster CM, Carr CE, Soukas AA. Biochemical and high throughput microscopic assessment of fat mass in Caenorhabditis elegans. J Vis Exp. 2013 doi: 10.3791/50180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–23. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, et al. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics. 2009;10:213. doi: 10.1186/1471-2164-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerstein MB, et al. Comparative analysis of the transcriptome across distant species. Nature. 2014;512:445–8. doi: 10.1038/nature13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 40.Deline ML, Vrablik TL, Watts JL. Dietary supplementation of polyunsaturated fatty acids in Caenorhabditis elegans. J Vis Exp. 2013 doi: 10.3791/50879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data are available at Sequence Read Archive (SRA) under accession number PRJNA343151. The code for RNA-seq analysis is available on the Brunet Lab Github: https://github.com/brunetlab.