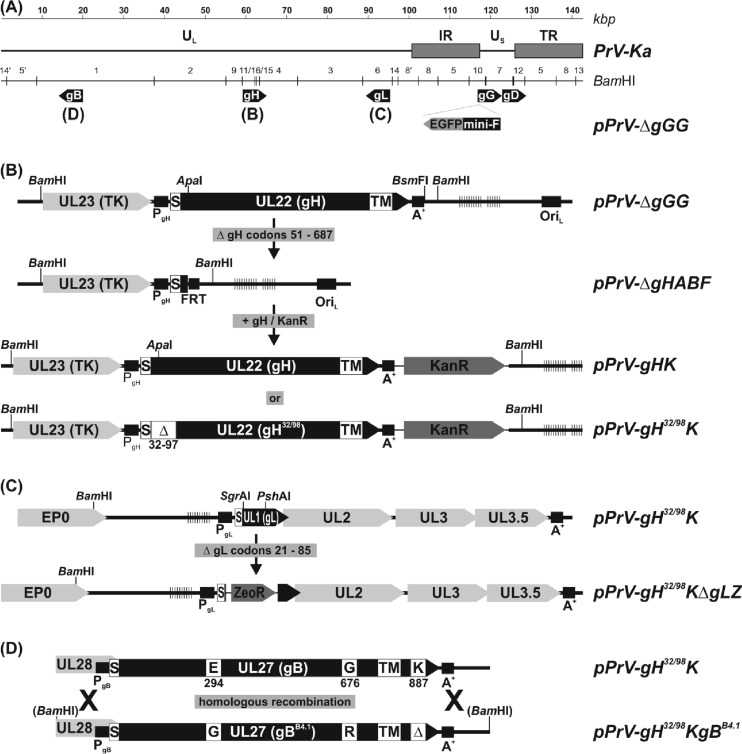
FIG 1.
Construction of virus mutants. (A) The wild-type PrV-Ka genome consists of unique long and short regions (UL and US, respectively), and inverted repeat sequences (IR, internal repeat; TR, terminal repeat). The positions of the relevant glycoprotein genes (gB, gD, gG, gH, and gL) are indicated. An EGFP expression cassette and a mini-F vector for BAC replication were inserted at the nonessential gG locus. (B) After deletion of the wild-type gH gene, modified forms were reinserted together with a kanamycin resistance gene (Kanr) by mutagenesis in E. coli. (C) In a similar manner, the gL gene was replaced by a zeocin resistance gene (Zeor). (D) Replacement of wild-type gB by mutated gBB4.1 was achieved by homologous recombination in BAC- and transfer plasmid-cotransfected RK13 cells and subsequent plaque purification of viable progeny viruses. Designations of virus recombinants, relevant restriction sites, regulatory sequences like promoters (PgX), polyadenylation signals (A+), an origin of DNA replication (ORIL), an Flp recombinase target site (FRT), and open reading frames (pointed rectangles) are indicated. The locations of signal peptides (S), transmembrane domains (TM), deletions (Δ), and amino acid substitutions in the deduced glycoproteins are highlighted. Vertical lines indicate tandem repeat sequences. TK, thymidine kinase. EP0, early protein 0.